Two-Fold Gene Expression Changes ( Down Regulated) in Mcf10ca1a
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Wide Analysis of 5-Hmc in the Peripheral Blood of Systemic Lupus Erythematosus Patients Using an Hmedip-Chip
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 35: 1467-1479, 2015 Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip WEIGUO SUI1*, QIUPEI TAN1*, MING YANG1, QIANG YAN1, HUA LIN1, MINGLIN OU1, WEN XUE1, JIEJING CHEN1, TONGXIANG ZOU1, HUANYUN JING1, LI GUO1, CUIHUI CAO1, YUFENG SUN1, ZHENZHEN CUI1 and YONG DAI2 1Guangxi Key Laboratory of Metabolic Diseases Research, Central Laboratory of Guilin 181st Hospital, Guilin, Guangxi 541002; 2Clinical Medical Research Center, the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital), Shenzhen, Guangdong 518020, P.R. China Received July 9, 2014; Accepted February 27, 2015 DOI: 10.3892/ijmm.2015.2149 Abstract. Systemic lupus erythematosus (SLE) is a chronic, Introduction potentially fatal systemic autoimmune disease characterized by the production of autoantibodies against a wide range Systemic lupus erythematosus (SLE) is a typical systemic auto- of self-antigens. To investigate the role of the 5-hmC DNA immune disease, involving diffuse connective tissues (1) and modification with regard to the onset of SLE, we compared is characterized by immune inflammation. SLE has a complex the levels 5-hmC between SLE patients and normal controls. pathogenesis (2), involving genetic, immunologic and envi- Whole blood was obtained from patients, and genomic DNA ronmental factors. Thus, it may result in damage to multiple was extracted. Using the hMeDIP-chip analysis and valida- tissues and organs, especially the kidneys (3). SLE arises from tion by quantitative RT-PCR (RT-qPCR), we identified the a combination of heritable and environmental influences. differentially hydroxymethylated regions that are associated Epigenetics, the study of changes in gene expression with SLE. -

Acetyl Group Coordinated Progression Through the Catalytic Cycle of an Arylalkylamine N-Acetyltransferase
RESEARCH ARTICLE Acetyl group coordinated progression through the catalytic cycle of an arylalkylamine N-acetyltransferase Adam A. Aboalroub, Ashleigh B. Bachman, Ziming Zhang, Dimitra Keramisanou, David J. Merkler, Ioannis Gelis* Department of Chemistry, University of South Florida, Tampa, Florida, United States of America * [email protected] a1111111111 a1111111111 a1111111111 Abstract a1111111111 a1111111111 The transfer of an acetyl group from acetyl-CoA to an acceptor amine is a ubiquitous bio- chemical transformation catalyzed by Gcn5-related N-acetyltransferases (GNATs). Although it is established that the reaction proceeds through a sequential ordered mecha- nism, the role of the acetyl group in driving the ordered formation of binary and ternary com- OPEN ACCESS plexes remains elusive. Herein, we show that CoA and acetyl-CoA alter the conformation of the substrate binding site of an arylalkylamine N-acetyltransferase (AANAT) to facilitate Citation: Aboalroub AA, Bachman AB, Zhang Z, Keramisanou D, Merkler DJ, Gelis I (2017) Acetyl interaction with acceptor substrates. However, it is the presence of the acetyl group within group coordinated progression through the the catalytic funnel that triggers high affinity binding. Acetyl group occupancy is relayed catalytic cycle of an arylalkylamine N- through a conserved salt bridge between the P-loop and the acceptor binding site, and is acetyltransferase. PLoS ONE 12(5): e0177270. manifested as differential dynamics in the CoA and acetyl-CoA-bound states. The capacity https://doi.org/10.1371/journal.pone.0177270 of the acetyl group carried by an acceptor to promote its tight binding even in the absence of Editor: Viswanathan V. Krishnan, California State CoA, but also its mutually exclusive position to the acetyl group of acetyl-CoA underscore its University Fresno, UNITED STATES importance in coordinating the progression of the catalytic cycle. -

Cytoplasmic Activation-Induced Cytidine Deaminase (AID) Exists in Stoichiometric Complex with Translation Elongation Factor 1Α (Eef1a)
Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1α (eEF1A) Julien Häsler, Cristina Rada, and Michael S. Neuberger1 Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom Edited by Frederick W. Alt, Howard Hughes Medical Institute, Harvard Medical School, Children’s Hospital Immune Disease Institute, Boston, MA, and approved October 12, 2011 (received for review April 27, 2011) Activation-induced cytidine deaminase (AID) is a B lymphocyte- results reveal that endogenous cytoplasmic AID partakes in a specific DNA deaminase that acts on the Ig loci to trigger antibody complex containing stoichiometric quantities of translation elon- gene diversification. Most AID, however, is retained in the cyto- gation factor 1α (eEF1A), with this association likely implicated in plasm and its nuclear abundance is carefully regulated because the regulation of AID’s intracellular trafficking. off-target action of AID leads to cancer. The nature of the cytosolic AID complex and the mechanisms regulating its release from the Results cytoplasm and import into the nucleus remain unknown. Here, we Flag-Tagging the Endogenous AID Locus in DT40 Cells. We generated show that cytosolic AID in DT40 B cells is part of an 11S complex derivatives of the DT40 B-cell line in which the endogenous AID and, using an endogenously tagged AID protein to avoid overex- locus was modified so as to incorporate a single Flag tag at the pression artifacts, that it is bound in good stoichiometry to the AID N terminus. To allow targeting of both alleles, one targeting translation elongation factor 1 alpha (eEF1A). -

Multiple Activities of Arl1 Gtpase in the Trans-Golgi Network Chia-Jung Yu1,2 and Fang-Jen S
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1691-1699 doi:10.1242/jcs.201319 COMMENTARY Multiple activities of Arl1 GTPase in the trans-Golgi network Chia-Jung Yu1,2 and Fang-Jen S. Lee3,4,* ABSTRACT typical features of an Arf-family GTPase, including an amphipathic ADP-ribosylation factors (Arfs) and ADP-ribosylation factor-like N-terminal helix and a consensus site for N-myristoylation (Lu et al., proteins (Arls) are highly conserved small GTPases that function 2001; Price et al., 2005). In yeast, recruitment of Arl1 to the Golgi as main regulators of vesicular trafficking and cytoskeletal complex requires a second Arf-like GTPase, Arl3 (Behnia et al., reorganization. Arl1, the first identified member of the large Arl family, 2004; Setty et al., 2003). Yeast Arl3 lacks a myristoylation site and is an important regulator of Golgi complex structure and function in is, instead, N-terminally acetylated; this modification is required for organisms ranging from yeast to mammals. Together with its effectors, its recruitment to the Golgi complex by Sys1. In mammalian cells, Arl1 has been shown to be involved in several cellular processes, ADP-ribosylation-factor-related protein 1 (Arfrp1), a mammalian including endosomal trans-Golgi network and secretory trafficking, lipid ortholog of yeast Arl3, plays a pivotal role in the recruitment of Arl1 droplet and salivary granule formation, innate immunity and neuronal to the trans-Golgi network (TGN) (Behnia et al., 2004; Panic et al., development, stress tolerance, as well as the response of the unfolded 2003b; Setty et al., 2003; Zahn et al., 2006). -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony
Supplementary Data Th2 and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in UBIOPRED Chih-Hsi Scott Kuo1.2, Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony Rowe3, Iaonnis Pandis2, Ana Sousa4, Julie Corfield5, Ratko Djukanovic6, Rene 7 7 8 2 1† Lutter , Peter J. Sterk , Charles Auffray , Yike Guo , Ian M. Adcock & Kian Fan 1†* # Chung on behalf of the U-BIOPRED consortium project team 1Airways Disease, National Heart & Lung Institute, Imperial College London, & Biomedical Research Unit, Biomedical Research Unit, Royal Brompton & Harefield NHS Trust, London, United Kingdom; 2Department of Computing & Data Science Institute, Imperial College London, United Kingdom; 3Janssen Research and Development, High Wycombe, Buckinghamshire, United Kingdom; 4Respiratory Therapeutic Unit, GSK, Stockley Park, United Kingdom; 5AstraZeneca R&D Molndal, Sweden and Areteva R&D, Nottingham, United Kingdom; 6Faculty of Medicine, Southampton University, Southampton, United Kingdom; 7Faculty of Medicine, University of Amsterdam, Amsterdam, Netherlands; 8European Institute for Systems Biology and Medicine, CNRS-ENS-UCBL, Université de Lyon, France. †Contributed equally #Consortium project team members are listed under Supplementary 1 Materials *To whom correspondence should be addressed: [email protected] 2 List of the U-BIOPRED Consortium project team members Uruj Hoda & Christos Rossios, Airways Disease, National Heart & Lung Institute, Imperial College London, UK & Biomedical Research Unit, Biomedical Research Unit, Royal -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

1 Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental
Page 1 of 255 Diabetes Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy Oliver J. Freeman1,2, Richard D. Unwin2,3, Andrew W. Dowsey2,3, Paul Begley2,3, Sumia Ali1, Katherine A. Hollywood2,3, Nitin Rustogi2,3, Rasmus S. Petersen1, Warwick B. Dunn2,3†, Garth J.S. Cooper2,3,4,5* & Natalie J. Gardiner1* 1 Faculty of Life Sciences, University of Manchester, UK 2 Centre for Advanced Discovery and Experimental Therapeutics (CADET), Central Manchester University Hospitals NHS Foundation Trust, Manchester Academic Health Sciences Centre, Manchester, UK 3 Centre for Endocrinology and Diabetes, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, UK 4 School of Biological Sciences, University of Auckland, New Zealand 5 Department of Pharmacology, Medical Sciences Division, University of Oxford, UK † Present address: School of Biosciences, University of Birmingham, UK *Joint corresponding authors: Natalie J. Gardiner and Garth J.S. Cooper Email: [email protected]; [email protected] Address: University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, United Kingdom Telephone: +44 161 275 5768; +44 161 701 0240 Word count: 4,490 Number of tables: 1, Number of figures: 6 Running title: Metabolic dysfunction in diabetic neuropathy 1 Diabetes Publish Ahead of Print, published online October 15, 2015 Diabetes Page 2 of 255 Abstract High glucose levels in the peripheral nervous system (PNS) have been implicated in the pathogenesis of diabetic neuropathy (DN). However our understanding of the molecular mechanisms which cause the marked distal pathology is incomplete. Here we performed a comprehensive, system-wide analysis of the PNS of a rodent model of DN. -
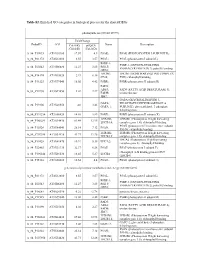
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4