Contig Protein Description Symbol Anterior Posterior Ratio
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table 1. Identified Proteins with Expression Significantly Altered in the Hippocampus of Rats of Exposed Group (Pb) Vs
Table 1. Identified proteins with expression significantly altered in the hippocampus of rats of exposed group (Pb) vs. Control. Fold Change Accession Id a Protein Description Score Pb P35213 14-3-3 protein beta/alpha 85420 −0.835 P62260 14-3-3 protein epsilon 96570 −0.878 P68511 14-3-3 protein eta 85420 −0.844 P68255 14-3-3 protein theta 85420 −0.835 P63102 14-3-3 protein zeta/delta 105051 −0.803 P13233 2',3'-cyclic-nucleotide 3'-phosphodiesterase 151400 1.405 P68035 Actin, alpha cardiac muscle 1 442584 −0.942 P68136 Actin, alpha skeletal muscle 441060 −0.970 P62738 Actin, aortic smooth muscle 438270 −0.970 P60711 Actin, cytoplasmic 1 630104 −0.942 P63259 Actin, cytoplasmic 2 630104 −0.942 P63269 Actin, gamma-enteric smooth muscle 438270 −0.951 Q05962 ADP/ATP translocase 1 60100 −0.554 Q09073 ADP/ATP translocase 2 49102 −0.482 P84079 ADP-ribosylation factor 1 34675 −0.644 P84082 ADP-ribosylation factor 2 22412 −0.644 P61206 ADP-ribosylation factor 3 34675 −0.619 P61751 ADP-ribosylation factor 4 22412 −0.670 P84083 ADP-ribosylation factor 5 22412 −0.625 P04764 Alpha-enolase 46219 −0.951 P23565 Alpha-internexin 9478 1.062 P37377 Alpha-synuclein 89619 −0.771 P13221 Aspartate aminotransferase, cytoplasmic 23661 1.083 P00507 Aspartate aminotransferase, mitochondrial 46049 1.116 P10719 ATP synthase subunit beta, mitochondrial 232442 −0.835 P85969 Beta-soluble NSF attachment protein 9638 1.419 Q63754 Beta-synuclein 66842 −0.779 P11275 Calcium/calmodulin-dependent protein kinase type II subunit alpha 181954 1.105 P08413 Calcium/calmodulin-dependent protein kinase type II subunit beta 80840 1.127 P15791 Calcium/calmodulin-dependent protein kinase type II subunit delta 62682 1.105 Int. -

Adaptive Laboratory Evolution Enhances Methanol Tolerance and Conversion in Engineered Corynebacterium Glutamicum
ARTICLE https://doi.org/10.1038/s42003-020-0954-9 OPEN Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum Yu Wang 1, Liwen Fan1,2, Philibert Tuyishime1, Jiao Liu1, Kun Zhang1,3, Ning Gao1,3, Zhihui Zhang1,3, ✉ ✉ 1234567890():,; Xiaomeng Ni1, Jinhui Feng1, Qianqian Yuan1, Hongwu Ma1, Ping Zheng1,2,3 , Jibin Sun1,3 & Yanhe Ma1 Synthetic methylotrophy has recently been intensively studied to achieve methanol-based biomanufacturing of fuels and chemicals. However, attempts to engineer platform micro- organisms to utilize methanol mainly focus on enzyme and pathway engineering. Herein, we enhanced methanol bioconversion of synthetic methylotrophs by improving cellular tolerance to methanol. A previously engineered methanol-dependent Corynebacterium glutamicum is subjected to adaptive laboratory evolution with elevated methanol content. Unexpectedly, the evolved strain not only tolerates higher concentrations of methanol but also shows improved growth and methanol utilization. Transcriptome analysis suggests increased methanol con- centrations rebalance methylotrophic metabolism by down-regulating glycolysis and up- regulating amino acid biosynthesis, oxidative phosphorylation, ribosome biosynthesis, and parts of TCA cycle. Mutations in the O-acetyl-L-homoserine sulfhydrylase Cgl0653 catalyzing formation of L-methionine analog from methanol and methanol-induced membrane-bound transporter Cgl0833 are proven crucial for methanol tolerance. This study demonstrates the importance of -

Regulation of Xenobiotic and Bile Acid Metabolism by the Anti-Aging Intervention Calorie Restriction in Mice
REGULATION OF XENOBIOTIC AND BILE ACID METABOLISM BY THE ANTI-AGING INTERVENTION CALORIE RESTRICTION IN MICE By Zidong Fu Submitted to the Graduate Degree Program in Pharmacology, Toxicology, and Therapeutics and the Graduate Faculty of the University of Kansas in partial fulfillment of the requirements for the degree of Doctor of Philosophy. Dissertation Committee ________________________________ Chairperson: Curtis Klaassen, Ph.D. ________________________________ Udayan Apte, Ph.D. ________________________________ Wen-Xing Ding, Ph.D. ________________________________ Thomas Pazdernik, Ph.D. ________________________________ Hao Zhu, Ph.D. Date Defended: 04-11-2013 The Dissertation Committee for Zidong Fu certifies that this is the approved version of the following dissertation: REGULATION OF XENOBIOTIC AND BILE ACID METABOLISM BY THE ANTI-AGING INTERVENTION CALORIE RESTRICTION IN MICE ________________________________ Chairperson: Curtis Klaassen, Ph.D. Date approved: 04-11-2013 ii ABSTRACT Calorie restriction (CR), defined as reduced calorie intake without causing malnutrition, is the best-known intervention to increase life span and slow aging-related diseases in various species. However, current knowledge on the exact mechanisms of aging and how CR exerts its anti-aging effects is still inadequate. The detoxification theory of aging proposes that the up-regulation of xenobiotic processing genes (XPGs) involved in phase-I and phase-II xenobiotic metabolism as well as transport, which renders a wide spectrum of detoxification, is a longevity mechanism. Interestingly, bile acids (BAs), the metabolites of cholesterol, have recently been connected with longevity. Thus, this dissertation aimed to determine the regulation of xenobiotic and BA metabolism by the well-known anti-aging intervention CR. First, the mRNA expression of XPGs in liver during aging was investigated. -
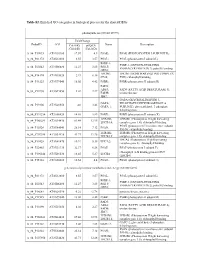
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Biosynthèse De Nouveaux Dérivés De L'alpha-Bisabolol Par Une Approche
THÈSE En vue de l’obtention du DOCTORAT DE L’UNIVERSITÉ DE TOULOUSE Délivré par l'Institut National des Sciences Appliquées de Toulouse Présentée et soutenue par Arthur SARRADE-LOUCHEUR Le 30 juin 2020 Biosynthèse de nouveaux dérivés de l'α-bisabolol par une approche de biologie synthèse Ecole doctorale : SEVAB - Sciences Ecologiques, Vétérinaires, Agronomiques et Bioingenieries Spécialité : Ingénieries microbienne et enzymatique Unité de recherche : TBI - Toulouse Biotechnology Institute, Bio & Chemical Engineering Thèse dirigée par Gilles TRUAN et Magali REMAUD-SIMEON Jury Mme Véronique DE BERARDINIS, Rapporteure, Chercheur CEA M. Jean-Etienne BASSARD, Rapporteur, Chargé de Recherche, CNRS Mme Danièle WERCK-REICHHART, Examinatrice, Directeur de Recherche Emérite, CNRS M. Gilles TRUAN, Directeur de thèse, Directeur de Recherche, CNRS Mme Magali REMAUD-SIMEON, Co-directrice de thèse, Professeur des Universités, INSA Mme Fayza DABOUSSI, Présidente, Directeur de Recherche INRAE 2 Author: Arthur Sarrade-Loucheur Year: 2020 Biosynthesis of new α-bisabolol derivatives through a synthetic biology approach The rise of synthetic biology now enables the production of new to nature molecules. In the frame of this thesis we focused on the diversification of the (+)-epi-α-bisabolol scaffold. This molecule coming from the plant Lippia dulcis belongs to the vast family of sesquiterpenes. While sesquiterpenes possess diverse biological activities, (+)-epi-α- bisabolol is the precursor of hernandulcin, an intense sweetener. However, the last oxidative step(s) of the hernandulcin biosynthetic pathway remain elusive. Rather than seeking the native oxidase responsible for hernandulcin synthesis among L. dulcis enzymes we turned our choice towards oxidative enzymes known to be promiscuous and that could functionalize (+)-epi-α-bisabolol in order to i) generate diversity from (+)-epi-α-bisabolol; ii) hopefully identify an oxidative enzyme catalyzing hernandulcin synthesis. -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing -

Serum Albumin OS=Homo Sapiens
Protein Name Cluster of Glial fibrillary acidic protein OS=Homo sapiens GN=GFAP PE=1 SV=1 (P14136) Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 Cluster of Isoform 3 of Plectin OS=Homo sapiens GN=PLEC (Q15149-3) Cluster of Hemoglobin subunit beta OS=Homo sapiens GN=HBB PE=1 SV=2 (P68871) Vimentin OS=Homo sapiens GN=VIM PE=1 SV=4 Cluster of Tubulin beta-3 chain OS=Homo sapiens GN=TUBB3 PE=1 SV=2 (Q13509) Cluster of Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 (P60709) Cluster of Tubulin alpha-1B chain OS=Homo sapiens GN=TUBA1B PE=1 SV=1 (P68363) Cluster of Isoform 2 of Spectrin alpha chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTAN1 (Q13813-2) Hemoglobin subunit alpha OS=Homo sapiens GN=HBA1 PE=1 SV=2 Cluster of Spectrin beta chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTBN1 PE=1 SV=2 (Q01082) Cluster of Pyruvate kinase isozymes M1/M2 OS=Homo sapiens GN=PKM PE=1 SV=4 (P14618) Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 Clathrin heavy chain 1 OS=Homo sapiens GN=CLTC PE=1 SV=5 Filamin-A OS=Homo sapiens GN=FLNA PE=1 SV=4 Cytoplasmic dynein 1 heavy chain 1 OS=Homo sapiens GN=DYNC1H1 PE=1 SV=5 Cluster of ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide OS=Homo sapiens GN=ATP1A2 PE=3 SV=1 (B1AKY9) Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 Dihydropyrimidinase-related protein 2 OS=Homo sapiens GN=DPYSL2 PE=1 SV=1 Cluster of Alpha-actinin-1 OS=Homo sapiens GN=ACTN1 PE=1 SV=2 (P12814) 60 kDa heat shock protein, mitochondrial OS=Homo -

Saturated Long-Chain Fatty Acid-Producing Bacteria Contribute
Zhao et al. Microbiome (2018) 6:107 https://doi.org/10.1186/s40168-018-0492-6 RESEARCH Open Access Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats Ling Zhao1†, Yufen Huang2†, Lin Lu1†, Wei Yang1, Tao Huang1, Zesi Lin3, Chengyuan Lin1,4, Hiuyee Kwan1, Hoi Leong Xavier Wong1, Yang Chen5, Silong Sun2, Xuefeng Xie2, Xiaodong Fang2,5, Huanming Yang6, Jian Wang6, Lixin Zhu7* and Zhaoxiang Bian1* Abstract Background: The gut microbiota is closely associated with gastrointestinal (GI) motility disorder, but the mechanism(s) by which bacteria interact with and affect host GI motility remains unclear. In this study, through using metabolomic and metagenomic analyses, an animal model of neonatal maternal separation (NMS) characterized by accelerated colonic motility and gut dysbiosis was used to investigate the mechanism underlying microbiota-driven motility dysfunction. Results: An excess of intracolonic saturated long-chain fatty acids (SLCFAs) was associated with enhanced bowel motility in NMS rats. Heptadecanoic acid (C17:0) and stearic acid (C18:0), as the most abundant odd- and even- numbered carbon SLCFAs in the colon lumen, can promote rat colonic muscle contraction and increase stool frequency. Increase of SLCFAs was positively correlated with elevated abundances of Prevotella, Lactobacillus, and Alistipes. Functional annotation found that the level of bacterial LCFA biosynthesis was highly enriched in NMS group. Essential synthetic genes Fabs were largely identified from the genera Prevotella, Lactobacillus, and Alistipes. Pseudo germ-free (GF) rats receiving fecal microbiota from NMS donors exhibited increased defecation frequency and upregulated bacterial production of intracolonic SLCFAs. Modulation of gut dysbiosis by neomycin effectively attenuated GI motility and reduced bacterial SLCFA generation in the colon lumen of NMS rats. -

Crystal Structure of Fabz-ACP Complex Reveals a Dynamic Seesaw-Like Catalytic Mechanism of Dehydratase in Fatty Acid Biosynthesis
Cell Research (2016) 26:1330-1344. ORIGINAL ARTICLE www.nature.com/cr Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis Lin Zhang1, 2, Jianfeng Xiao3, Jianrong Xu1, Tianran Fu1, Zhiwei Cao1, Liang Zhu1, 2, Hong-Zhuan Chen1, 2, Xu Shen3, Hualiang Jiang3, Liang Zhang1, 2 1Department of Pharmacology and Chemical Biology, Shanghai Jiao Tong University School of Medicine, Shanghai, China; 2Shanghai Universities Collaborative Innovation Center for Translational Medicine, Shanghai, China; 3State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China Fatty acid biosynthesis (FAS) is a vital process in cells. Fatty acids are essential for cell assembly and cellular me- tabolism. Abnormal FAS directly correlates with cell growth delay and human diseases, such as metabolic syndromes and various cancers. The FAS system utilizes an acyl carrier protein (ACP) as a transporter to stabilize and shuttle the growing fatty acid chain throughout enzymatic modules for stepwise catalysis. Studying the interactions between enzymatic modules and ACP is, therefore, critical for understanding the biological function of the FAS system. How- ever, the information remains unclear due to the high flexibility of ACP and its weak interaction with enzymatic mod- ules. We present here a 2.55 Å crystal structure of type II FAS dehydratase FabZ in complex with holo-ACP, which exhibits a highly symmetrical FabZ hexamer-ACP3 stoichiometry with each ACP binding to a FabZ dimer subunit. Further structural analysis, together with biophysical and computational results, reveals a novel dynamic seesaw-like ACP binding and catalysis mechanism for the dehydratase module in the FAS system, which is regulated by a critical gatekeeper residue (Tyr100 in FabZ) that manipulates the movements of the β-sheet layer. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

Structure of L-Rhamnose Isomerase in Complex with L
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Open Bio 3 (2013) 35–40 journal homepage: www.elsevier.com/locate/febsopenbio Structure of l-rhamnose isomerase in complex with l-rhamnopyranose demonstrates the sugar-ring opening mechanism and the role of a substrate sub-binding site Hiromi Yoshidaa, Akihide Yoshiharab, Misa Teraokaa, Satoshi Yamashitaa, Ken Izumorib, Shigehiro Kamitoria,* aLife Science Research Center and Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan bRare Sugar Research Center and Faculty of Agriculture, Kagawa University, Miki-cho, Kagawa 761-0795, Japan article info abstract Article history: L-Rhamnose isomerase (L-RhI) catalyzes the reversible isomerization of L-rhamnose to L-rhamnulose. Received 27 November 2012 Previously determined X-ray structures of L-RhI showed a hydride-shift mechanism for the isomeriza- Accepted 30 November 2012 tion of substrates in a linear form, but the mechanism for opening of the sugar-ring is still unclear. To elucidate this mechanism, we determined X-ray structures of a mutant L-RhI in complex with L- Keywords: rhamnopyranose and D-allopyranose. Results suggest that a catalytic water molecule, which acts as an l-Rhamnose isomerase acid/base catalyst in the isomerization reaction, is likely to be involved in pyranose-ring opening, and Pseudomonas stutzeri Sugar-ring opening mechanism that a newly found substrate sub-binding site in the vicinity of the catalytic site may recognize different Rare sugar anomers of substrates. X-ray structure C 2012 The Authors.