Supplemental Methods Patients and Controls. Coronary Arteries from KD
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Acetyl Group Coordinated Progression Through the Catalytic Cycle of an Arylalkylamine N-Acetyltransferase
RESEARCH ARTICLE Acetyl group coordinated progression through the catalytic cycle of an arylalkylamine N-acetyltransferase Adam A. Aboalroub, Ashleigh B. Bachman, Ziming Zhang, Dimitra Keramisanou, David J. Merkler, Ioannis Gelis* Department of Chemistry, University of South Florida, Tampa, Florida, United States of America * [email protected] a1111111111 a1111111111 a1111111111 Abstract a1111111111 a1111111111 The transfer of an acetyl group from acetyl-CoA to an acceptor amine is a ubiquitous bio- chemical transformation catalyzed by Gcn5-related N-acetyltransferases (GNATs). Although it is established that the reaction proceeds through a sequential ordered mecha- nism, the role of the acetyl group in driving the ordered formation of binary and ternary com- OPEN ACCESS plexes remains elusive. Herein, we show that CoA and acetyl-CoA alter the conformation of the substrate binding site of an arylalkylamine N-acetyltransferase (AANAT) to facilitate Citation: Aboalroub AA, Bachman AB, Zhang Z, Keramisanou D, Merkler DJ, Gelis I (2017) Acetyl interaction with acceptor substrates. However, it is the presence of the acetyl group within group coordinated progression through the the catalytic funnel that triggers high affinity binding. Acetyl group occupancy is relayed catalytic cycle of an arylalkylamine N- through a conserved salt bridge between the P-loop and the acceptor binding site, and is acetyltransferase. PLoS ONE 12(5): e0177270. manifested as differential dynamics in the CoA and acetyl-CoA-bound states. The capacity https://doi.org/10.1371/journal.pone.0177270 of the acetyl group carried by an acceptor to promote its tight binding even in the absence of Editor: Viswanathan V. Krishnan, California State CoA, but also its mutually exclusive position to the acetyl group of acetyl-CoA underscore its University Fresno, UNITED STATES importance in coordinating the progression of the catalytic cycle. -

Linc-DYNC2H1-4 Promotes EMT and CSC Phenotypes by Acting As a Sponge of Mir-145 in Pancreatic Cancer Cells
Citation: Cell Death and Disease (2017) 8, e2924; doi:10.1038/cddis.2017.311 OPEN Official journal of the Cell Death Differentiation Association www.nature.com/cddis Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells Yuran Gao1, Zhicheng Zhang2,3, Kai Li1,3, Liying Gong1, Qingzhu Yang1, Xuemei Huang1, Chengcheng Hong1, Mingfeng Ding*,2 and Huanjie Yang*,1 The acquisition of epithelial–mesenchymal transition (EMT) and/or existence of a sub-population of cancer stem-like cells (CSC) are associated with malignant behavior and chemoresistance. To identify which factor could promote EMT and CSC formation and uncover the mechanistic role of such factor is important for novel and targeted therapies. In the present study, we found that the long intergenic non-coding RNA linc-DYNC2H1-4 was upregulated in pancreatic cancer cell line BxPC-3-Gem with acquired gemcitabine resistance. Knockdown of linc-DYNC2H1-4 decreased the invasive behavior of BxPC-3-Gem cells while ectopic expression of linc-DYNC2H1-4 promoted the acquisition of EMT and stemness of the parental sensitive cells. Linc-DYNC2H1-4 upregulated ZEB1, the EMT key player, which led to upregulation and downregulation of its targets vimentin and E-cadherin respectively, as well as enhanced the expressions of CSC makers Lin28, Nanog, Sox2 and Oct4. Linc-DYNC2H1-4 is mainly located in the cytosol. Mechanically, it could sponge miR-145 that targets ZEB1, Lin28, Nanog, Sox2, Oct4 to restore these EMT and CSC-associated genes expressions. We proved that MMP3, the nearby gene of linc-DYNC2H1-4 in the sense strand, was also a target of miR-145. -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Identification and Interaction Analysis of Molecular Markers in Pancreatic
medRxiv preprint doi: https://doi.org/10.1101/2020.12.20.20248601; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification and Interaction Analysis of Molecular Markers in Pancreatic Ductal Adenocarcinoma by Integrated Bioinformatics Analysis and Molecular Docking Experiments Basavaraj Vastrad1 , Chanabasayya Vastrad *2, Anandkumar Tengli3 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics,Chanabasava Nilaya, Bharthinagar, Dharwad, Karanataka 580001, India. 3. Department of Pharmaceutical Chemistry, JSS College of Pharmacy, Mysuru and JSS Academy of Higher Education & Research, Mysuru- 570015, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.20.20248601; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract The current investigation aimed to mine therapeutic molecular targets that play an key part in the advancement of pancreatic ductal adenocarcinoma (PDAC). The expression profiling by high throughput sequencing dataset profile GSE133684 dataset was downloaded from the Gene Expression Omnibus (GEO) database. -

Comparative Transcriptome Analysis of Embryo Invasion in the Mink Uterus
Placenta 75 (2019) 16–22 Contents lists available at ScienceDirect Placenta journal homepage: www.elsevier.com/locate/placenta Comparative transcriptome analysis of embryo invasion in the mink uterus T ∗ Xinyan Caoa,b, , Chao Xua,b, Yufei Zhanga,b, Haijun Weia,b, Yong Liuc, Junguo Caoa,b, Weigang Zhaoa,b, Kun Baoa,b, Qiong Wua,b a Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China b State Key Laboratory for Molecular Biology of Special Economic Animal and Plant Science, Chinese Academy of Agricultural Sciences, Changchun, China c Key Laboratory of Embryo Development and Reproductive Regulation of Anhui Province, College of Biological and Food Engineering, Fuyang Teachers College, Fuyang, China ABSTRACT Introduction: In mink, as many as 65% of embryos die during gestation. The causes and the mechanisms of embryonic mortality remain unclear. The purpose of our study was to examine global gene expression changes during embryo invasion in mink, and thereby to identify potential signaling pathways involved in implantation failure and early pregnancy loss. Methods: Illumina's next-generation sequencing technology (RNA-Seq) was used to analyze the differentially expressed genes (DEGs) in implantation (IMs) and inter- implantation sites (inter-IMs) of uterine tissue. Results: We identified a total of 606 DEGs, including 420 up- and 186 down-regulated genes in IMs compared to inter-IMs. Gene annotation analysis indicated multiple biological pathways to be significantly enriched for DEGs, including immune response, ECM complex, cytokine activity, chemokine activity andprotein binding. The KEGG pathway including cytokine-cytokine receptor interaction, Jak-STAT, TNF and the chemokine signaling pathway were the most enriched. -

Supplemental Table 1. Complete Gene Lists and GO Terms from Figure 3C
Supplemental Table 1. Complete gene lists and GO terms from Figure 3C. Path 1 Genes: RP11-34P13.15, RP4-758J18.10, VWA1, CHD5, AZIN2, FOXO6, RP11-403I13.8, ARHGAP30, RGS4, LRRN2, RASSF5, SERTAD4, GJC2, RHOU, REEP1, FOXI3, SH3RF3, COL4A4, ZDHHC23, FGFR3, PPP2R2C, CTD-2031P19.4, RNF182, GRM4, PRR15, DGKI, CHMP4C, CALB1, SPAG1, KLF4, ENG, RET, GDF10, ADAMTS14, SPOCK2, MBL1P, ADAM8, LRP4-AS1, CARNS1, DGAT2, CRYAB, AP000783.1, OPCML, PLEKHG6, GDF3, EMP1, RASSF9, FAM101A, STON2, GREM1, ACTC1, CORO2B, FURIN, WFIKKN1, BAIAP3, TMC5, HS3ST4, ZFHX3, NLRP1, RASD1, CACNG4, EMILIN2, L3MBTL4, KLHL14, HMSD, RP11-849I19.1, SALL3, GADD45B, KANK3, CTC- 526N19.1, ZNF888, MMP9, BMP7, PIK3IP1, MCHR1, SYTL5, CAMK2N1, PINK1, ID3, PTPRU, MANEAL, MCOLN3, LRRC8C, NTNG1, KCNC4, RP11, 430C7.5, C1orf95, ID2-AS1, ID2, GDF7, KCNG3, RGPD8, PSD4, CCDC74B, BMPR2, KAT2B, LINC00693, ZNF654, FILIP1L, SH3TC1, CPEB2, NPFFR2, TRPC3, RP11-752L20.3, FAM198B, TLL1, CDH9, PDZD2, CHSY3, GALNT10, FOXQ1, ATXN1, ID4, COL11A2, CNR1, GTF2IP4, FZD1, PAX5, RP11-35N6.1, UNC5B, NKX1-2, FAM196A, EBF3, PRRG4, LRP4, SYT7, PLBD1, GRASP, ALX1, HIP1R, LPAR6, SLITRK6, C16orf89, RP11-491F9.1, MMP2, B3GNT9, NXPH3, TNRC6C-AS1, LDLRAD4, NOL4, SMAD7, HCN2, PDE4A, KANK2, SAMD1, EXOC3L2, IL11, EMILIN3, KCNB1, DOK5, EEF1A2, A4GALT, ADGRG2, ELF4, ABCD1 Term Count % PValue Genes regulation of pathway-restricted GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, SMAD protein phosphorylation 9 6.34 1.31E-08 ENG pathway-restricted SMAD protein GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, phosphorylation -
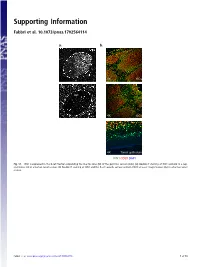
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

A Functional Study of ADAMTS7 Gene Variants
A functional study of ADAMTS7 gene variants Xiangyuan Pu A thesis submitted to the University of London for the degree of Doctor of Philosophy Centre for Clinical Pharmacology William Harvey Research Institute Barts and the London School of Medicine and Dentistry Queen Mary, University of London March, 2014 I dedicate this thesis to my parents and my fiancée. Without their endless love, support and encouragement, none of my achievements would be possible. 2 Abstract Background: Recent studies have revealed an association between genetic variants at the ADAMTS7 (a disintegrin-like and metalloprotease with thrombospondin type 1 motif, 7) locus and susceptibility to coronary artery disease (CAD). ADAMTS-7 has been reported to facilitate vascular smooth muscle cell (VSMC) migration and promote neointima formation. We sought to study the functional mechanisms underlying this relationship and to further investigate the role of ADAMTS-7 in atherosclerosis. Methods and Results: In vitro assays showed that the CAD-associated non-synonymous single nucleotide polymorphism rs3825807, which results in a serine to proline (Ser-to- Pro) substitution at residue 214 in the ADAMTS-7 pro-domain, affected ADAMTS-7 pro- domain cleavage. Immunohistochemical analyses showed that ADAMTS-7 localised to vascular smooth muscle cells (VSMCs) and endothelial cells (ECs) in human coronary and carotid atherosclerotic plaques. Cell migration assays demonstrated that VSMCs and ECs from individuals who were homozygous for the adenine (A) allele (encoding the Ser214 isoform) had increased migratory ability compared with cells from individuals who were homozygous for the G allele (encoding the Pro214 isoform). Western blot analyses revealed that media conditioned by VSMCs of the A/A genotype contained more cleaved ADAMTS-7 pro-domain and more of the cleaved form of thrombospondin-5 (TSP-5, an ADAMTS-7 substrate that had been shown to be produced by VSMCs and inhibit VSMC migration). -

Effect of Nanoparticles on the Expression and Activity of Matrix Metalloproteinases
Nanotechnol Rev 2018; 7(6): 541–553 Review Magdalena Matysiak-Kucharek*, Magdalena Czajka, Krzysztof Sawicki, Marcin Kruszewski and Lucyna Kapka-Skrzypczak Effect of nanoparticles on the expression and activity of matrix metalloproteinases https://doi.org/10.1515/ntrev-2018-0110 Received September 14, 2018; accepted October 11, 2018; previously 1 Introduction published online November 15, 2018 Matrix metallopeptidases, commonly known as matrix Abstract: Matrix metallopeptidases, commonly known metalloproteinases (MMPs), are zinc-dependent proteo- as matrix metalloproteinases (MMPs), are a group of pro- lytic enzymes whose primary function is the degradation teolytic enzymes whose main function is the remodeling and remodeling of extracellular matrix (ECM) compo- of the extracellular matrix. Changes in the activity of nents. ECM is a complex, dynamic structure that condi- these enzymes are observed in many pathological states, tions the proper tissue architecture. MMPs by digesting including cancer metastases. An increasing body of evi- ECM proteins eliminate structural barriers and allow dence indicates that nanoparticles (NPs) can lead to the cell migration. Moreover, by hydrolyzing extracellularly deregulation of MMP expression and/or activity both in released proteins, MMPs can change the activity of many vitro and in vivo. In this work, we summarized the current signal peptides, such as growth factors, cytokines, and state of knowledge on the impact of NPs on MMPs. The chemokines. MMPs are involved in many physiological literature analysis showed that the impact of NPs on MMP processes, such as embryogenesis, reproduction cycle, or expression and/or activity is inconclusive. NPs exhibit wound healing; however, their increased activity is also both stimulating and inhibitory effects, which might be associated with a number of pathological conditions, such dependent on multiple factors, such as NP size and coat- as diabetes, cardiovascular diseases and neurodegenera- ing or a cellular model used in the research.