WO 2016/004043 Al O O©
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Acetyl Group Coordinated Progression Through the Catalytic Cycle of an Arylalkylamine N-Acetyltransferase
RESEARCH ARTICLE Acetyl group coordinated progression through the catalytic cycle of an arylalkylamine N-acetyltransferase Adam A. Aboalroub, Ashleigh B. Bachman, Ziming Zhang, Dimitra Keramisanou, David J. Merkler, Ioannis Gelis* Department of Chemistry, University of South Florida, Tampa, Florida, United States of America * [email protected] a1111111111 a1111111111 a1111111111 Abstract a1111111111 a1111111111 The transfer of an acetyl group from acetyl-CoA to an acceptor amine is a ubiquitous bio- chemical transformation catalyzed by Gcn5-related N-acetyltransferases (GNATs). Although it is established that the reaction proceeds through a sequential ordered mecha- nism, the role of the acetyl group in driving the ordered formation of binary and ternary com- OPEN ACCESS plexes remains elusive. Herein, we show that CoA and acetyl-CoA alter the conformation of the substrate binding site of an arylalkylamine N-acetyltransferase (AANAT) to facilitate Citation: Aboalroub AA, Bachman AB, Zhang Z, Keramisanou D, Merkler DJ, Gelis I (2017) Acetyl interaction with acceptor substrates. However, it is the presence of the acetyl group within group coordinated progression through the the catalytic funnel that triggers high affinity binding. Acetyl group occupancy is relayed catalytic cycle of an arylalkylamine N- through a conserved salt bridge between the P-loop and the acceptor binding site, and is acetyltransferase. PLoS ONE 12(5): e0177270. manifested as differential dynamics in the CoA and acetyl-CoA-bound states. The capacity https://doi.org/10.1371/journal.pone.0177270 of the acetyl group carried by an acceptor to promote its tight binding even in the absence of Editor: Viswanathan V. Krishnan, California State CoA, but also its mutually exclusive position to the acetyl group of acetyl-CoA underscore its University Fresno, UNITED STATES importance in coordinating the progression of the catalytic cycle. -

Potential High-Impact Interventions Report Priority Area 02: Cancer
AHRQ Healthcare Horizon Scanning System – Potential High-Impact Interventions Report Priority Area 02: Cancer Prepared for: Agency for Healthcare Research and Quality U.S. Department of Health and Human Services 540 Gaither Road Rockville, MD 20850 www.ahrq.gov Contract No. HHSA290201000006C Prepared by: ECRI Institute 5200 Butler Pike Plymouth Meeting, PA 19462 December 2012 Statement of Funding and Purpose This report incorporates data collected during implementation of the Agency for Healthcare Research and Quality (AHRQ) Healthcare Horizon Scanning System by ECRI Institute under contract to AHRQ, Rockville, MD (Contract No. HHSA290201000006C). The findings and conclusions in this document are those of the authors, who are responsible for its content, and do not necessarily represent the views of AHRQ. No statement in this report should be construed as an official position of AHRQ or of the U.S. Department of Health and Human Services. This report’s content should not be construed as either endorsements or rejections of specific interventions. As topics are entered into the System, individual topic profiles are developed for technologies and programs that appear to be close to diffusion into practice in the United States. Those reports are sent to various experts with clinical, health systems, health administration, and/or research backgrounds for comment and opinions about potential for impact. The comments and opinions received are then considered and synthesized by ECRI Institute to identify interventions that experts deemed, through the comment process, to have potential for high impact. Please see the methods section for more details about this process. This report is produced twice annually and topics included may change depending on expert comments received on interventions issued for comment during the preceding 6 months. -

Investigator Initiated Study IRB-29839 an Open-Label Pilot Study To
Investigator Initiated Study IRB-29839 An open-label pilot study to evaluate the efficacy and safety of a combination treatment of Sonidegib and BKM120 for the treatment of advanced basal cell carcinomas Version 05 September 2016 NCT02303041 DATE: 12Dec2018 1 Coordinating Center Stanford Cancer Center 875 Blake Wilbur Drive Stanford, CA 94305 And 450 Broadway, MC 5334 Redwood City, CA 94603 Protocol Director and Principal Investigator Anne Lynn S Chang, MD, Director of Dermatological Clinical Trials 450 Broadway St, MC 5334 Redwood City, CA 94603 [email protected] Co-Investigator Anthony Oro, MD PhD 450 Broadway St, MC 5334 Redwood City, CA 94603 [email protected] Biostatistician Shufeng Li, MS 450 Broadway St, MC 5334 Redwood City, CA 94603 [email protected] Study Coordinator Ann Moffat 450 Broadway St, MC 5334 Redwood City, CA 94603 [email protected] 2 Table of Contents 1 Background ................................................................. 7 1.1 Disease Background ..................................................... 7 1.2 Hedgehog Pathway and mechanism of action ............................... 7 1.3 PI3K Pathway and mechanism of action ................................... 9 1.4 Sonidegib Compound Information ............ Error! Bookmark not defined. 1.4.1 Preclinical Studies for Sonidegib ....................................................................11 1.4.2 Muscular system...............................................................................................13 1.4.3 Skeletal system ................................................................................................13 -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -

Genomic Oncology: Moving Beyond the Tip of the Iceberg Jane De Lartigue, Phd
FeatureCommunity Report Genomic oncology: moving beyond the tip of the iceberg Jane de Lartigue, PhD istorically, cancer has been diagnosed and in patients with lung cancer, even the most efec- treated on the basis of the tissue of ori- tive targeted therapies can fail if used in the wrong Hgin. Te promise of personalized therapy, patient population.5,6 matched more precisely to an individual’s tumor, In recognition of this issue, the oncology feld has was ushered in with the development of molecularly developed molecular biomarkers that can predict targeted therapies, based on a greater understanding response, or lack thereof, to targeted therapy. Drugs of cancer as a genomic-driven disease. Here, we dis- are now commonly being evaluated in trials that cuss some of the evolution of genomic oncology, the select eligible patients on the basis of biomarker pos- inherent complexities and challenges, and how novel itivity, and a number of companion diagnostics have clinical trial designs are among the strategies being been codeveloped to assist in these eforts (Table 1). developed to address them and shape the future of Notable successes include the development of the personalized medicine in cancer. monoclonal antibody trastuzumab for patients with breast cancers that have human epidermal growth The evolution of genomic oncology factor receptor 2 (HER2) gene amplifcation or In the 15 years since the frst map of the human HER2 protein overexpression,7 and the small mol- genome emerged, genetics has become an inte- ecule BRAF kinase inhibitor -

Meeting Key Synthetic Challenges in Amanitin Synthesis with a New Cytotoxic Analog: 50-Hydroxy- Cite This: Chem
Chemical Science View Article Online EDGE ARTICLE View Journal | View Issue Meeting key synthetic challenges in amanitin synthesis with a new cytotoxic analog: 50-hydroxy- Cite this: Chem. Sci., 2020, 11, 11927 0 † All publication charges for this article 6 -deoxy-amanitin have been paid for by the Royal Society of Chemistry Alla Pryyma, Kaveh Matinkhoo, Antonio A. W. L. Wong and David M. Perrin * Appreciating the need to access synthetic analogs of amanitin, here we report the synthesis of 50-hydroxy- Received 29th July 2020 60-deoxy-amanitin, a novel, rationally-designed bioactive analog and constitutional isomer of a-amanitin, Accepted 2nd October 2020 that is anticipated to be used as a payload for antibody drug conjugates. In completing this synthesis, we DOI: 10.1039/d0sc04150e meet the challenge of diastereoselective sulfoxidation by presenting two high-yielding and rsc.li/chemical-science diastereoselective sulfoxidation approaches to afford the more toxic (R)-sulfoxide. drug-tolerant cell subpopulations.7 Examples include ADCs for Creative Commons Attribution-NonCommercial 3.0 Unported Licence. Introduction targeting human epidermal growth factor receptor 2 and pros- 8 9 a-Amanitin, the deadliest of the amatoxins produced by the tate specic membrane antigen. With but a few exceptions, death-cap mushroom Amanita phalloides, is a potent, orally nearly all bioconjugates to a cytotoxic amanitin have emerged 1 a available inhibitor of RNA polymerase II (pol II) (Ki 10 nM), from naturally-sourced -amanitin. To date, conjugation that has been validated as a payload for targeted cancer handles used for a-amanitin-based bioconjugates include the d- 2a therapy.2 First described in 1907 3 and isolated in 1941,4 a- hydroxyl of (2S,3R,4R)-4,5-dihydroxyisoleucine (DHIle), the 10 0 amanitin is a compact bicyclic octapeptide that has been asparagine side chain, and the 6 -hydroxyl of the tryptathio- 11 indispensable for probing RNA pol II-catalysed transcription in nine staple. -

NINDS Custom Collection II
ACACETIN ACEBUTOLOL HYDROCHLORIDE ACECLIDINE HYDROCHLORIDE ACEMETACIN ACETAMINOPHEN ACETAMINOSALOL ACETANILIDE ACETARSOL ACETAZOLAMIDE ACETOHYDROXAMIC ACID ACETRIAZOIC ACID ACETYL TYROSINE ETHYL ESTER ACETYLCARNITINE ACETYLCHOLINE ACETYLCYSTEINE ACETYLGLUCOSAMINE ACETYLGLUTAMIC ACID ACETYL-L-LEUCINE ACETYLPHENYLALANINE ACETYLSEROTONIN ACETYLTRYPTOPHAN ACEXAMIC ACID ACIVICIN ACLACINOMYCIN A1 ACONITINE ACRIFLAVINIUM HYDROCHLORIDE ACRISORCIN ACTINONIN ACYCLOVIR ADENOSINE PHOSPHATE ADENOSINE ADRENALINE BITARTRATE AESCULIN AJMALINE AKLAVINE HYDROCHLORIDE ALANYL-dl-LEUCINE ALANYL-dl-PHENYLALANINE ALAPROCLATE ALBENDAZOLE ALBUTEROL ALEXIDINE HYDROCHLORIDE ALLANTOIN ALLOPURINOL ALMOTRIPTAN ALOIN ALPRENOLOL ALTRETAMINE ALVERINE CITRATE AMANTADINE HYDROCHLORIDE AMBROXOL HYDROCHLORIDE AMCINONIDE AMIKACIN SULFATE AMILORIDE HYDROCHLORIDE 3-AMINOBENZAMIDE gamma-AMINOBUTYRIC ACID AMINOCAPROIC ACID N- (2-AMINOETHYL)-4-CHLOROBENZAMIDE (RO-16-6491) AMINOGLUTETHIMIDE AMINOHIPPURIC ACID AMINOHYDROXYBUTYRIC ACID AMINOLEVULINIC ACID HYDROCHLORIDE AMINOPHENAZONE 3-AMINOPROPANESULPHONIC ACID AMINOPYRIDINE 9-AMINO-1,2,3,4-TETRAHYDROACRIDINE HYDROCHLORIDE AMINOTHIAZOLE AMIODARONE HYDROCHLORIDE AMIPRILOSE AMITRIPTYLINE HYDROCHLORIDE AMLODIPINE BESYLATE AMODIAQUINE DIHYDROCHLORIDE AMOXEPINE AMOXICILLIN AMPICILLIN SODIUM AMPROLIUM AMRINONE AMYGDALIN ANABASAMINE HYDROCHLORIDE ANABASINE HYDROCHLORIDE ANCITABINE HYDROCHLORIDE ANDROSTERONE SODIUM SULFATE ANIRACETAM ANISINDIONE ANISODAMINE ANISOMYCIN ANTAZOLINE PHOSPHATE ANTHRALIN ANTIMYCIN A (A1 shown) ANTIPYRINE APHYLLIC -

Tanibirumab (CUI C3490677) Add to Cart
5/17/2018 NCI Metathesaurus Contains Exact Match Begins With Name Code Property Relationship Source ALL Advanced Search NCIm Version: 201706 Version 2.8 (using LexEVS 6.5) Home | NCIt Hierarchy | Sources | Help Suggest changes to this concept Tanibirumab (CUI C3490677) Add to Cart Table of Contents Terms & Properties Synonym Details Relationships By Source Terms & Properties Concept Unique Identifier (CUI): C3490677 NCI Thesaurus Code: C102877 (see NCI Thesaurus info) Semantic Type: Immunologic Factor Semantic Type: Amino Acid, Peptide, or Protein Semantic Type: Pharmacologic Substance NCIt Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor tyrosine kinase expressed by endothelial cells, while VEGF is overexpressed in many tumors and is correlated to tumor progression. PDQ Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor -
Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Anamika Kothari Quang Ong Ron Caspi An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Peter D Karp Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Csac1394711Cyc: Candidatus Saccharibacteria bacterium RAAC3_TM7_1 Cellular Overview Connections between pathways are omitted for legibility. Tim Holland TM7C00001G0420 TM7C00001G0109 TM7C00001G0953 TM7C00001G0666 TM7C00001G0203 TM7C00001G0886 TM7C00001G0113 TM7C00001G0247 TM7C00001G0735 TM7C00001G0001 TM7C00001G0509 TM7C00001G0264 TM7C00001G0176 TM7C00001G0342 TM7C00001G0055 TM7C00001G0120 TM7C00001G0642 TM7C00001G0837 TM7C00001G0101 TM7C00001G0559 TM7C00001G0810 TM7C00001G0656 TM7C00001G0180 TM7C00001G0742 TM7C00001G0128 TM7C00001G0831 TM7C00001G0517 TM7C00001G0238 TM7C00001G0079 TM7C00001G0111 TM7C00001G0961 TM7C00001G0743 TM7C00001G0893 TM7C00001G0630 TM7C00001G0360 TM7C00001G0616 TM7C00001G0162 TM7C00001G0006 TM7C00001G0365 TM7C00001G0596 TM7C00001G0141 TM7C00001G0689 TM7C00001G0273 TM7C00001G0126 TM7C00001G0717 TM7C00001G0110 TM7C00001G0278 TM7C00001G0734 TM7C00001G0444 TM7C00001G0019 TM7C00001G0381 TM7C00001G0874 TM7C00001G0318 TM7C00001G0451 TM7C00001G0306 TM7C00001G0928 TM7C00001G0622 TM7C00001G0150 TM7C00001G0439 TM7C00001G0233 TM7C00001G0462 TM7C00001G0421 TM7C00001G0220 TM7C00001G0276 TM7C00001G0054 TM7C00001G0419 TM7C00001G0252 TM7C00001G0592 TM7C00001G0628 TM7C00001G0200 TM7C00001G0709 TM7C00001G0025 TM7C00001G0846 TM7C00001G0163 TM7C00001G0142 TM7C00001G0895 TM7C00001G0930 Detoxification Carbohydrate Biosynthesis DNA combined with a 2'- di-trans,octa-cis a 2'- Amino Acid Degradation an L-methionyl- TM7C00001G0190 superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis (E. -

WSC 2000-01 Conference 1
The Armed Forces Institute of Pathology Department of Veterinary Pathology WEDNESDAY SLIDE CONFERENCE 2002-2003 CONFERENCE 17 12 February 2003 Conference Moderator: LTC Gary Zaucha, DVM Diplomate, ACVP, ABT, and ACVPM Chief, Department of Comparative Pathology Walter Reed Army Institute of Research Silver Spring, MD 20910 CASE I – 1614-1 (AFIP 2850109) Signalment: 5-month-old outbred female Swiss-Webster mouse History: Sentinel mouse in a laboratory colony housed on dirty bedding from other mouse cages, part of an infectious disease surveillance program. Euthanized due to lethargy and unthriftiness. Gross Pathology: Stomach distended approximately three times by gas, fluid, and partially digested food. Kidneys shrunken, pale, and pitted. Multifocal hemorrhagic necrosis and thrombosis of the ovaries bilaterally. Laboratory Results: Negative for all murine infectious pathogens tested in the colony surveillance program by serology, respiratory and intestinal cultures, fecal examination, anal tape, and skin scrapings. Contributor’s Morphologic Diagnosis: Kidney: Chronic nephropathy characterized by membranous glomerulopathy, lymphoplasmacytic adventitial vasculitis and perivasculitis, tubular degeneration, ectasia, and regeneration with protein, hemoglobin, cellular, waxy, and granular casts, and lymphoplasmacytic and histiocytic interstitial nephritis, severe. Contributor’s Comment: This case is consistent with the previously reported syndrome of gastric dilatation and chronic nephropathy in mice exposed to dirty bedding [1]. Although the mean age of affected mice in the published report was 10 months, similar lesions are sometimes found in animals as young as 3 or 4 months of age. The kidney disease appears immune-mediated and is presumably the result of chronic high antigen exposure, although there may be more than one inciting process. -
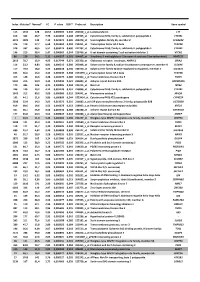
Index Mutated* Normal* FC P Value FDR** Probe Set Description Gene Symbol
Index Mutated* Normal* FC P value FDR** Probe set Description Gene symbol 113 1342 128 10,52 0,000503 0,240 202018_s_at Lactotransferrin LTF 616 362 46,7 7,76 0,003493 0,308 237395_at Cytochrome P450, family 4, subfamily Z, polypeptide 1 CYP4Z1 3073 1009 142 7,10 0,021674 0,385 206378_at Secretoglobin, family 2A, member 2 SCGB2A2 376 119 17,7 6,68 0,001962 0,284 214451_at Transcription factor AP-2 beta TFAP2B 578 337 60,5 5,57 0,003174 0,300 227702_at Cytochrome P450, family 4, subfamily X, polypeptide 1 CYP4X1 146 210 38,4 5,47 0,000669 0,244 219768_at V-set domain containing T cell activation inhibitor 1 VTCN1 86 129 24,2 5,31 0,000327 0,201 204607_at 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) HMGCS2 2613 78,7 15,9 4,95 0,017744 0,371 205358_at Glutamate receptor, ionotropic, AMPA 2 GRIA2 116 23,2 4,83 4,81 0,000513 0,240 203908_at Solute carrier family 4, sodium bicarbonate cotransporter, member 4 SLC4A4 117 79,4 18,0 4,42 0,000518 0,240 239723_at Solute carrier family 40 (iron-regulated transporter), member 1 SLC40A1 625 56,6 13,0 4,34 0,003549 0,308 1553394_a_atTranscription factor AP-2 beta TFAP2B 413 148 34,6 4,28 0,002175 0,286 240304_s_at Transmembrane channel-like 5 TMC5 5181 216 50,9 4,25 0,039946 0,421 223864_at Ankyrin repeat domain 30A ANKRD30A 179 446 106 4,21 0,000826 0,244 223315_at Netrin 4 NTN4 246 120 29,2 4,12 0,001128 0,251 210096_at Cytochrome P450, family 4, subfamily B, polypeptide 1 CYP4B1 1043 312 80,2 3,89 0,006080 0,319 204041_at Monoamine oxidase B MAOB 192 44,1 11,6 3,80 0,000899 0,244 -

Evaluation of the Allometric Exponents in Prediction of Human Drug Clearance
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2014 Evaluation of the Allometric Exponents in Prediction of Human Drug Clearance Da Zhang Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Other Pharmacy and Pharmaceutical Sciences Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/3533 This Dissertation is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. ©Da Zhang, 2014 All Rights Reserve EVALUATION OF THE ALLOMETRIC EXPONENTS IN PREDICTION OF HUMAN DRUG CLEARANCE A Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy at Virginia Commonwealth University By Da Zhang Master of Science, University of Arizona, 2004 Director: F. Douglas Boudinot, Ph.D. Professor, School of Pharmacy, VCU Virginia Commonwealth University Richmond, Virginia August, 2014 ACKNOWLEDGEMENTS First and foremost, I would like to sincerely thank my advisor, Dr. F. Douglas Boudinot, for giving me the opportunity to pursue graduate studies under his guidance and for his continuous professional support, guidance, encouragement and patience throughout my graduate program. He was always delighted in sharing his vast knowledge and kind warmth. I would also like to thank Dr. Ahmad for his scientific advice and support through the complet ion of my degree. He has been a kind and helpful mentor for me. I would like to acknowledge my graduate committee members, Drs.