Suppl. Fig. 1 A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Novel Nucleobase-Ascorbate Transporter
bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Identification of a novel Nucleobase-Ascorbate 2 Transporter family member in fish and amphibians 3 Diogo Oliveira*,1, André M. Machado*,1, Tiago Cardoso1, Mónica Lopes-Marques1, L. Filipe 4 C. Castro1,2● and Raquel Ruivo1● 5 1CIIMAR – Interdisciplinary Centre of Marine and Environmental Research, U. Porto – University 6 of Porto, Porto, Portugal 7 2Department of Biology, Faculty of Sciences, U. Porto - University of Porto, Portugal 8 *Equal contribution 9 ●Corresponding authors at: CIIMAR, Terminal de Cruzeiros do Porto de Leixões, Av. General 10 Norton de Matos s/n, 4450-208 Matosinhos, Portugal. Tel.: +351 223 401 831 11 12 Running title: Novel uric acid transporter in fish and amphibians 13 14 15 bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 16 Abstract:Nucleobase-Ascorbate Transporter (NAT) family includes ascorbic acid, nucleobases 17 and uric acid transporters: with a broad evolutionary distribution. In vertebrates, four members 18 have been previously recognized, the ascorbate transporters Slc23a1 and Slc3a2, the nucleobase 19 transporter Slc23a4 and an orphan transporter Slc23a3. -

Glutamate, Aspartate and Nucleotide Transporters in the SLC17 Family Form Four Main Phylogenetic Clusters: Evolution and Tissue
Sreedharan et al. BMC Genomics 2010, 11:17 http://www.biomedcentral.com/1471-2164/11/17 RESEARCH ARTICLE Open Access Glutamate, aspartate and nucleotide transporters in the SLC17 family form four main phylogenetic clusters: evolution and tissue expression Smitha Sreedharan1, Jafar HA Shaik1, Pawel K Olszewski1,2, Allen S Levine2,3, Helgi B Schiöth1, Robert Fredriksson1* Abstract Background: The SLC17 family of transporters transports the amino acids: glutamate and aspartate, and, as shown recently, also nucleotides. Vesicular glutamate transporters are found in distinct species, such as C. elegans, but the evolutionary origin of most of the genes in this family has been obscure. Results: Our phylogenetic analysis shows that the SLC17 family consists of four main phylogenetic clades which were all present before the divergence of the insect lineage. One of these clades has not been previously described and it is not found in vertebrates. The clade containing Slc17a9 had the most restricted evolutionary history with only one member in most species. We detected expression of Slc17a1-17a4 only in the peripheral tissues but not in the CNS, while Slc17a5- Slc17a9 are highly expressed in both the CNS and periphery. Conclusions: The in situ hybridization studies on vesicular nucleotide transporter revealed high expression throughout the cerebral cortex, certain areas in the hippocampus and in specific nuclei of the hypothalamus and thalamus. Some of the regions with high expression, such as the medial habenula and the dentate gyrus of the hippocampus, are important sites for purinergic neurotransmission. Noteworthy, other areas relying on purine- mediated signaling, such as the molecular layer of the dentate gyrus and the periaqueductal gray, lack or have a very low expression of Slc17a9, suggesting that there could be another nucleotide transporter in these regions. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Modulation of Urate Transport by Drugs
pharmaceutics Review Modulation of Urate Transport by Drugs Péter Tátrai 1, Franciska Erd˝o 2, Gabriella Dörnyei 3 and Péter Krajcsi 1,2,3,* 1 Solvo Biotechnology, Science Park, Building B2, 4-20 Irinyi József utca, H-1117 Budapest, Hungary; [email protected] 2 Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, H-1083 Budapest, Hungary; [email protected] 3 Department of Morphology and Physiology, Faculty of Health Sciences, Semmelweis University, H-1088 Budapest, Hungary; [email protected] * Correspondence: [email protected] Abstract: Background: Serum urate (SU) levels in primates are extraordinarily high among mammals. Urate is a Janus-faced molecule that acts physiologically as a protective antioxidant but provokes inflammation and gout when it precipitates at high concentrations. Transporters play crucial roles in urate disposition, and drugs that interact with urate transporters either by intention or by accident may modulate SU levels. We examined whether in vitro transporter interaction studies may clarify and predict such effects. Methods: Transporter interaction profiles of clinically proven urate-lowering (uricosuric) and hyperuricemic drugs were compiled from the literature, and the predictive value of in vitro-derived cut-offs like Cmax/IC50 on the in vivo outcome (clinically relevant decrease or increase of SU) was assessed. Results: Interaction with the major reabsorptive urate transporter URAT1 appears to be dominant over interactions with secretory transporters in determining the net effect of a drug on SU levels. In vitro inhibition interpreted using the recommended cut-offs is useful at predicting the clinical outcome. Conclusions: In vitro safety assessments regarding urate transport should be done early in drug development to identify candidates at risk of causing major imbalances. -

Supporting Information
Supporting Information Table S1. List of confirmed SLC transporters represented in Canine GeneChip. SLC family Members detected Members not detected SLC1: The high affinity glutamate and neutral amino acid SLC1A1 SLC1A2, SLC1A3, SLC1A6 transporter family SLC2: The facilitative GLUT transporter family SLC2A1, SLC2A8 SLC2A3, SLC2A9 SLC3: The heavy subunits of the heteromeric amino acid SLC3A1 transporters SLC4: The bicarbonate transporter family SLC4A11 SLC4A4, SLC4A8 SLC5: The sodium glucose cotransporter family SLC5A6 SLC5A3, SLC5A10, SLC5A12 SLC6: The sodium- and chloride- dependent SLC6A6, SLC6A12 SLCA18 neurotransmitter transporter family SLC7: The cationic amino acid transporter/glycoprotein- NR associated family SLC8: The Na+/Ca2+ exchanger family SLC8A1 SLC9: The Na+/H+ exchanger family SLC9A1, SLC9A6, SLC9A9 SLC10: The sodium bile salt cotransport family SLC10A2 SLC11: The proton coupled metal ion transporter family NR SLC12: The electroneutral cation-Cl cotransporter family SLC12A3, SLC12A6, SLC12A8 SLC13: The human Na+-sulfate/carboxylate cotransporter SLC13A2 family SLC14: The urea transporter family NR SLC15: The proton oligopeptide cotransporter family SLC15A2, SLC15A4 SLC15A1 SLC16: The monocarboxylate transporter family SLC16A13 SLC16A4 SLC17: The vesicular glutamate transporter family SLC17A3, SLC17A7 SLC18: The vesicular amine transporter family NR SLC19: The folate/thiamine transporter family NR SLC20: The type III Na+-phosphate cotransporter family NR SLC21/SLCO: The organic anion transporting family SLC21A3, SLC21A8, -

Perkinelmer Genomics to Request the Saliva Swab Collection Kit for Patients That Cannot Provide a Blood Sample As Whole Blood Is the Preferred Sample
Autism and Intellectual Disability TRIO Panel Test Code TR002 Test Summary This test analyzes 2429 genes that have been associated with Autism and Intellectual Disability and/or disorders associated with Autism and Intellectual Disability with the analysis being performed as a TRIO Turn-Around-Time (TAT)* 3 - 5 weeks Acceptable Sample Types Whole Blood (EDTA) (Preferred sample type) DNA, Isolated Dried Blood Spots Saliva Acceptable Billing Types Self (patient) Payment Institutional Billing Commercial Insurance Indications for Testing Comprehensive test for patients with intellectual disability or global developmental delays (Moeschler et al 2014 PMID: 25157020). Comprehensive test for individuals with multiple congenital anomalies (Miller et al. 2010 PMID 20466091). Patients with autism/autism spectrum disorders (ASDs). Suspected autosomal recessive condition due to close familial relations Previously negative karyotyping and/or chromosomal microarray results. Test Description This panel analyzes 2429 genes that have been associated with Autism and ID and/or disorders associated with Autism and ID. Both sequencing and deletion/duplication (CNV) analysis will be performed on the coding regions of all genes included (unless otherwise marked). All analysis is performed utilizing Next Generation Sequencing (NGS) technology. CNV analysis is designed to detect the majority of deletions and duplications of three exons or greater in size. Smaller CNV events may also be detected and reported, but additional follow-up testing is recommended if a smaller CNV is suspected. All variants are classified according to ACMG guidelines. Condition Description Autism Spectrum Disorder (ASD) refers to a group of developmental disabilities that are typically associated with challenges of varying severity in the areas of social interaction, communication, and repetitive/restricted behaviors. -

RNA-Seq Reveals Conservation of Function Among the Yolk Sacs Of
RNA-seq reveals conservation of function among the PNAS PLUS yolk sacs of human, mouse, and chicken Tereza Cindrova-Daviesa, Eric Jauniauxb, Michael G. Elliota,c, Sungsam Gongd,e, Graham J. Burtona,1, and D. Stephen Charnock-Jonesa,d,e,1,2 aCentre for Trophoblast Research, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, United Kingdom; bElizabeth Garret Anderson Institute for Women’s Health, Faculty of Population Health Sciences, University College London, London, WC1E 6BT, United Kingdom; cSt. John’s College, University of Cambridge, Cambridge, CB2 1TP, United Kingdom; dDepartment of Obstetrics and Gynaecology, University of Cambridge, Cambridge, CB2 0SW, United Kingdom; and eNational Institute for Health Research, Cambridge Comprehensive Biomedical Research Centre, Cambridge, CB2 0QQ, United Kingdom Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved May 5, 2017 (received for review February 14, 2017) The yolk sac is phylogenetically the oldest of the extraembryonic yolk sac plays a critical role during organogenesis (3–5, 8–10), membranes. The human embryo retains a yolk sac, which goes there are limited data to support this claim. Obtaining experi- through primary and secondary phases of development, but its mental data for the human is impossible for ethical reasons, and importance is controversial. Although it is known to synthesize thus we adopted an alternative strategy. Here, we report RNA proteins, its transport functions are widely considered vestigial. sequencing (RNA-seq) data derived from human and murine yolk Here, we report RNA-sequencing (RNA-seq) data for the human sacs and compare them with published data from the yolk sac of and murine yolk sacs and compare those data with data for the the chicken. -
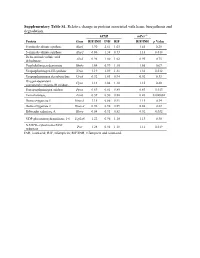
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator- -

Conserved Genes from Simon's Simplex Collection in Drosoph Human Gene HGNC Flybaseid Fly Gene A2M 7 Fbgn00
Supplemental Table 1: Conserved genes from Simon's Simplex Collection in Drosophila melanogaster Human Gene HGNC FlyBaseID Fly Gene A2M 7 FBgn0041181 Tep3 A2ML1 23336 FBgn0041180 Tep4 AASS 17366 FBgn0286198 LKRSDH ABCA1 29 FBgn0083956 CG34120 ABCA13 14638 FBgn0083956 CG34120 ABCA2 32 FBgn0031171 CG1801 ABCA7 37 FBgn0083956 CG34120 ABCA8 38 FBgn0034493 CG8908 ABCB6 47 FBgn0038376 Hmt-1 ABCC4 55 FBgn0038740 CG4562 ABCC5 56 FBgn0039644 rdog ABCG1 73 FBgn0020762 Atet ABCG2 74 FBgn0003996 w ABHD12 15868 FBgn0034419 CG15111 ABI2 24011 FBgn0020510 Abi ABL1 76 FBgn0000017 Abl ABL2 77 FBgn0000017 Abl ABR 81 FBgn0025836 RhoGAP1A ACACB 85 FBgn0033246 ACC ACE 2707 FBgn0012037 Ance ACHE 108 FBgn0000024 Ace ACP2 123 FBgn0000032 Acph-1 ACTN4 166 FBgn0000667 Actn ACTR6 24025 FBgn0011741 Arp6 ACTRT3 24022 FBgn0000045 Act79B ADAM18 196 FBgn0259110 mmd ADAMTS7 223 FBgn0029791 CG4096 ADAMTSL1 14632 FBgn0051619 nolo ADAMTSL4 19706 FBgn0032252 loh ADCY5 236 FBgn0263131 CG43373 ADD3 245 FBgn0263391 hts ADRBK2 290 FBgn0260798 Gprk1 AEBP2 24051 FBgn0086655 jing AGAP1 16922 FBgn0028509 CenG1A AGAP2 16921 FBgn0028509 CenG1A AGK 21869 FBgn0260750 Mulk AGO1 3262 FBgn0262739 AGO1 AGTRAP 13539 FBgn0052638 CG32638 AK1 361 FBgn0022709 Adk1 AKAP1 367 FBgn0263987 spoon AKAP9 379 FBgn0086690 Plp AKR1B15 37281 FBgn0086254 CG6084 AKR1C2 385 FBgn0086254 CG6084 AKR1D1 388 FBgn0086254 CG6084 AKT2 392 FBgn0010379 Akt1 ALDH18A1 9722 FBgn0037146 CG7470 ALDH1L1 3978 FBgn0032945 CG8665 ALDH3A1 405 FBgn0010548 Aldh-III ALDH5A1 408 FBgn0039349 Ssadh ALS2 443 FBgn0037116 Als2 AMPD2 469 -

Supplemental Data
Supplementary Table 1. Gene sets from Figure 6. Lists of genes from each individual gene set defined in Figure 6, including the fold-change in expression of each gene in treatment group pair-wise comparisons. ENSEMBL: Ensembl gene identifier; Symbol: official gene symbol; logFC: log fold change; p value: significance of fold-change in a pair-wise comparison, P<0.05 cut-off; FDR: false discovery rate, expected proportion of false positives among the differentially expressed genes in a pair-wise comparison (FDR<0.25 cut-off). Sup. Table 1 SET I CP versus Sal CP versus CP+DCA DCA versus Sal ENSEMBL Symbol logFC PValue FDR logFC PValue FDR logFC PValue FDR Desc ENSMUSG00000020326 Ccng1 2.64 0.00 0.00 -0.06 0.13 0.96 0.40 0.00 0.23 cyclin G1 [Source:MGI Symbol;Acc:MGI:102890] ENSMUSG00000031886 Ces2e 3.97 0.00 0.00 -0.24 0.02 0.28 0.01 1.00 1.00 carboxylesterase 2E [Source:MGI Symbol;Acc:MGI:2443170] ENSMUSG00000041959 S100a10 2.31 0.00 0.00 -0.21 0.02 0.23 -0.11 0.53 1.00 S100 calcium binding protein A10 (calpactin) [Source:MGI Symbol;Acc:MGI:1339468] ENSMUSG00000092341 Malat1 1.09 0.00 0.00 -0.11 0.20 1.00 0.66 0.00 0.00 metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) [Source:MGI Symbol;Acc:MGI:1919539] ENSMUSG00000072949 Acot1 1.73 0.00 0.00 -0.22 0.01 0.12 -0.44 0.01 1.00 acyl-CoA thioesterase 1 [Source:MGI Symbol;Acc:MGI:1349396] ENSMUSG00000064339 mt-Rnr2 1.09 0.00 0.00 -0.08 0.17 1.00 0.67 0.00 0.07 mitochondrially encoded 16S rRNA [Source:MGI Symbol;Acc:MGI:102492] ENSMUSG00000025934 Gsta3 1.86 0.00 0.00 -0.28 -

Table SI. List of Gout Candidate Genes. A1CF GCKR NIPAL1
Table SI. List of gout candidate genes. A1CF GCKR NIPAL1 SLC16A5 SLC2A1 ABCC4 GEF NLRP3 SLC16A6 SLC2A10 ABCG2 HADH NOD2 SLC16A7 SLC2A11 ACADS HIST1H2BF NRG4 SLC16A8 SLC2A12 ACVR1B HIST1H4E OSTC SLC16A9 SLC2A13 ACVR2A HLF P2RX7 SLC17A1 SLC2A14 ACVRL1 HNF1B P2X7R SLC17A2 SLC2A2 ADRB3 HNF4G PDK2 SLC17A3 SLC2A3 AGL HPRT1 PDZK1 SLC17A4 SLC2A3P1 ALDH16A1 IGF1R PFKM SLC17A5 SLC2A3P2 ALDH2 IGF1R PKLR SLC17A6 SLC2A3P4 ALDOB IL12B PLA2G12A SLC17A7 SLC2A4 ALPK1 IL18R1 PPARGC1A SLC17A8 SLC2A5 AMPD1 IL1A PPARGC1B SLC17A9 SLC2A6 AOX1 IL1B PRKAG2 SLC22A1 SLC2A7 ASAH2 IL23R PRKG2 SLC22A10 SLC2A8 ATXN2 IL33 PRPS1 SLC22A11 SLC2A9 B3GNT4 IL8 PRPSAP1 SLC22A12 SLC2AXP1 BAZ1B INHBB PTPN11 SLC22A13 SLC37A4 BCAS3 INHBC PYGM SLC22A14 STAT4 C17orf82 ITPR1 R3HDM2 SLC22A15 STC1 CARD15 JAK2 REN SLC22A16 TGFB1 CARD8 KCNQ1 RFX3 SLC22A17 TGM4 CD14 LEF1 RREB1 SLC22A18 TLR2 CLNK LRP2 SFMBT1 SLC22A2 TLR4 CNIH2 LRRC16A SGK1 SLC22A20 TMEM171 CNTN4 LTBP3 SGMS2 SLC22A23 TNXIP COMT MAF SLC16A1 SLC22A24 TRIM46 CPT2 MEFV SLC16A10 SLC22A25 UBE2Q2 CUX2 MLXIPL SLC16A11 SLC22A3 UGT1A CYP2U1 MOCOS SLC16A12 SLC22A31 UMOD DAPK1 MTHFR SLC16A13 SLC22A4 UOX DCP1A MUSTN1 SLC16A14 SLC22A5 VEGFA EGF MYD88 SLC16A14P1 SLC22A6 VEGFA ELOVL6 MYL2 SLC16A2 SLC22A7 WDR1 FAM35A NCF1 SLC16A3 SLC22A8 XDH G6PC NFAT5 SLC16A4 SLC22A9 ZNF518B Table SII. Results of conventional radiography, autoanti‑ bodies and HLA‑B27 tests of patients with gout in three gout pedigrees. Individual ID HLA‑B27 ANA+ENA Joint X‑ray F1_I:1 ‑ ‑ + F1_II:1 ‑ ‑ + F2_I:1 ‑ ‑ + F2_II:1 ‑ ‑ NA F2_II:7 ‑ ‑ ‑ F2_III:1 ‑ ‑ NA F2_III:9 ‑ ‑ ‑ F3_41 ‑ ‑ + F3_45 ‑ ‑ ‑ F3_ II:2 ‑ ‑ + F3_ III:2 ‑ ‑ NA F3_ III:3 ‑ ‑ NA Joint X‑ray ‘+’ means the patient has the typical manifestation of cortical erosion, soft tissue swelling, tophi or wormy appearance. -

In Silico Prioritization of Transporter-Drug Relationships From
bioRxiv preprint doi: https://doi.org/10.1101/381335; this version posted July 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 In silico prioritization of transporter-drug relationships from drug 2 sensitivity screens 3 4 Running title: Computational identification of SLC-drug associations 5 6 César-Razquin A.1, Girardi E.1, Yang M.2,3, Brehme M.3,5, Sáez-Rodríguez J.2,3,4, Superti-Furga 7 G.1,6* 8 9 1. CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, 10 Vienna, Austria 11 2. Heidelberg University, Heidelberg, Germany 12 3. Joint Research Centre for Computational Biomedicine (JRC-COMBINE), RWTH Aachen 13 University, Faculty of Medicine, Aachen, Germany 14 4. European Molecular Biology Laboratory - European Bioinformatics Institute, Wellcome 15 Genome Campus, Cambridge, UK 16 5. CBmed - Center for Biomarker Research in Medicine GmbH, Graz, Austria 17 6. Center for Physiology and Pharmacology, Medical University of Vienna, Vienna, Austria 18 19 * Correspondence: 20 Prof. Giulio Superti-Furga 21 [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/381335; this version posted July 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license.