Identification of a Novel Nucleobase-Ascorbate Transporter
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -
Suppl. Fig. 1 A
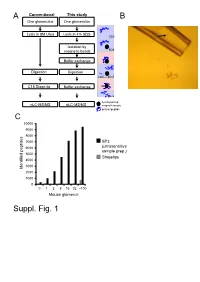
A Conventional This study B One glomerulus One glomerulus Lysis in 8M Urea Lysis in 4% SDS SDS Isolation by magnetic beads SDS Buffer exchange Digestion Digestion Try LysC pH>7 C18 Stage tip Buffer exchange pH<4 functionalized nLC-MS/MS nLC-MS/MS magnetic beads protein/peptide C 10000 9000 8000 7000 SP3 6000 (ultrasensitive sample prep.) 5000 Stagetips 4000 3000 Identified peptides 2000 1000 0 0 1 2 8 16 32 ~100 Mouse glomeruli Suppl. Fig. 1 S1. Sample preparation methods. A. Scheme of conventional (C18 Stage Tip) and ultrasensitive proteomic sample preparation methods. B. Manual microdissection and isolation of single glomeruli. The arrow indicates a single isolated glomerulus subjected to proteome analysis. The tip capacity is 10µl. C. Stagetips, a conventional proteomic sample preparation method, is compared with ultrasensitive (SP3) proteome analysis. The indicated amounts of mouse glomeruli were prepared by the respective sample preparation protocols, and peptide numbers after nLC-MS/MS are plotted (all FDR<0.01). A 100 1 Tubule B 80 60 40 Intensity CCD 20 40 0 100 20 S1 TAL 80 0 Tubule proximal tubule Loading... 60 0 Intensity 40 S2 20 -20 control Component 2 (22.5%) S3 0 0 10 20 30 40 50 60 -40 -20 0 20 40 Time (min) Component 2 (47.1%) C D 6000 Mouse proximal tubule 5000 8 4000 3000 7 (iBAQ) Loading... 10 2000 log 6 Number of peptides 1000 0 5 Control mouse mouse mouse human proximal mTAL CCD proximal 0 1500 tubule tubule Rank protein (S1) (S1) R=0.7 E F 8 ACTB Human proximal tubule 8 7 SLC9A3R1 ATP1A1 SLC3A2 SLC25A5 ATP1B1 7 SLC5A2 SLC25A3 CUBN CLTC SLC27A2 SLC5A12 LRP2 6 SLC25A13 SLC25A10 SLC5A1 SLC22A6 6 SLC25A4 Loading.. -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -

Glutamate, Aspartate and Nucleotide Transporters in the SLC17 Family Form Four Main Phylogenetic Clusters: Evolution and Tissue
Sreedharan et al. BMC Genomics 2010, 11:17 http://www.biomedcentral.com/1471-2164/11/17 RESEARCH ARTICLE Open Access Glutamate, aspartate and nucleotide transporters in the SLC17 family form four main phylogenetic clusters: evolution and tissue expression Smitha Sreedharan1, Jafar HA Shaik1, Pawel K Olszewski1,2, Allen S Levine2,3, Helgi B Schiöth1, Robert Fredriksson1* Abstract Background: The SLC17 family of transporters transports the amino acids: glutamate and aspartate, and, as shown recently, also nucleotides. Vesicular glutamate transporters are found in distinct species, such as C. elegans, but the evolutionary origin of most of the genes in this family has been obscure. Results: Our phylogenetic analysis shows that the SLC17 family consists of four main phylogenetic clades which were all present before the divergence of the insect lineage. One of these clades has not been previously described and it is not found in vertebrates. The clade containing Slc17a9 had the most restricted evolutionary history with only one member in most species. We detected expression of Slc17a1-17a4 only in the peripheral tissues but not in the CNS, while Slc17a5- Slc17a9 are highly expressed in both the CNS and periphery. Conclusions: The in situ hybridization studies on vesicular nucleotide transporter revealed high expression throughout the cerebral cortex, certain areas in the hippocampus and in specific nuclei of the hypothalamus and thalamus. Some of the regions with high expression, such as the medial habenula and the dentate gyrus of the hippocampus, are important sites for purinergic neurotransmission. Noteworthy, other areas relying on purine- mediated signaling, such as the molecular layer of the dentate gyrus and the periaqueductal gray, lack or have a very low expression of Slc17a9, suggesting that there could be another nucleotide transporter in these regions. -

The Genetic Landscape of the Human Solute Carrier (SLC) Transporter Superfamily
Human Genetics (2019) 138:1359–1377 https://doi.org/10.1007/s00439-019-02081-x ORIGINAL INVESTIGATION The genetic landscape of the human solute carrier (SLC) transporter superfamily Lena Schaller1 · Volker M. Lauschke1 Received: 4 August 2019 / Accepted: 26 October 2019 / Published online: 2 November 2019 © The Author(s) 2019 Abstract The human solute carrier (SLC) superfamily of transporters is comprised of over 400 membrane-bound proteins, and plays essential roles in a multitude of physiological and pharmacological processes. In addition, perturbation of SLC transporter function underlies numerous human diseases, which renders SLC transporters attractive drug targets. Common genetic polymorphisms in SLC genes have been associated with inter-individual diferences in drug efcacy and toxicity. However, despite their tremendous clinical relevance, epidemiological data of these variants are mostly derived from heterogeneous cohorts of small sample size and the genetic SLC landscape beyond these common variants has not been comprehensively assessed. In this study, we analyzed Next-Generation Sequencing data from 141,456 individuals from seven major human populations to evaluate genetic variability, its functional consequences, and ethnogeographic patterns across the entire SLC superfamily of transporters. Importantly, of the 204,287 exonic single-nucleotide variants (SNVs) which we identifed, 99.8% were present in less than 1% of analyzed alleles. Comprehensive computational analyses using 13 partially orthogonal algorithms that predict the functional impact of genetic variations based on sequence information, evolutionary conserva- tion, structural considerations, and functional genomics data revealed that each individual genome harbors 29.7 variants with putative functional efects, of which rare variants account for 18%. Inter-ethnic variability was found to be extensive, and 83% of deleterious SLC variants were only identifed in a single population. -

Supplemental Text and Figures
Supplemental Materials and Methods Experimental bone metastasis assay Primary PCa cells were sorted by GFP marker from mTmG+ tumors, and 105 cells in 20μL PBS were injected using Hamilton syringe into the tibia of 6-week old NSG mice. Mice were monitored biweekly for moribund signs for euthanasia and organ harvest. Noninvasive mouse and ex vivo imaging MRI imaging with Bruker ICON and fluorescence imaging of fresh organs with metastasis enumeration were recently described (Lu et al. 2017). Primary prostate cell sphere formation assay Isolate of primary cells from prostate, culture and counting of prostatospheres on Matrigel were performed as described (Lukacs et al. 2010). For organoid culture assay, we followed a published matrigel embedding method (Chua et al. 2014). RNA-Seq and differential gene expression Total RNA was isolated from prostate tumors using Direct-zol RNA MiniPrep Kit (Zymo Research) and processed for stranded total RNA-Seq using Illumina HiSeq 4000 at Sequencing and Microarray Facility at MD Anderson Cancer Center. The differential expression analysis was performed using the DESeq2 package of R. P-values obtained after multiple binomial tests were adjusted using BH method. Significant genes are defined by using a cut-off of 0.05 on the BH corrected p-value and an absolute log2 fold change value of at least 1.5. Histology and western blot H&E stain, immunohistochemical (IHC) and western blot were performed as previously described (Ding et al. 2011; Wang et al. 2016). Primary antibodies for IHC include Ki67 (Fisher, RM-9106-S1), cleaved caspase 3 (Cell Signaling Technology aka CST, 9661), cyclin D1 (Fisher, clone SP4), TGFBR2 (Abcam, ab61213), BMPR2 (Abcam, ab130206), AR (EMD Millipore, 06-680), phospho- Akt (CST, 4060), GFP (CST, 2956), E-Cadherin (CST, 14472). -

Genetic Variants in a Sodium-Dependent Vitamin C
Clinical science Br J Ophthalmol: first published as 10.1136/bjophthalmol-2018-312257 on 15 November 2018. Downloaded from Genetic variants in a sodium-dependent vitamin C transporter gene and age-related cataract Ravilla D Ravindran,1 Periasamy Sundaresan,2 Tiruvengada Krishnan,3 Praveen Vashist,4 Giovanni Maraini,5 Vijayan Saravanan,2 Usha Chakravarthy,6 Liam Smeeth,7 Dorothea Nitsch,7 Ian S Young,6 Astrid E Fletcher7 ► Additional material is ABSTRact in a rural population in India found no benefit in published online only. To view Background Cataract is a major health burden in the progression of opacities over a 5-year period please visit the journal online many countries and a significant problem in India. While from supplementation with high-dose vitamins A, (http:// dx. doi. org/ 10. 1136/ 5 bjophthalmol- 2018- 312257). observational studies show lower cataract risk with C and E. The lack of convincing evidence from increasing dietary or plasma vitamin C, randomised this and other RCTs is not surprising since a priori 1 Cataract Services, Aravind Eye controlled trials of supplements have been negative. it is unlikely that vitamin supplements given for a Hospital, Madurai, India Genetic variants in vitamin C transporter proteins relatively short duration would prevent an age-re- 2Department of Genetics, Dr.G.Venkataswamy Eye (SLC23A1), especially rs33972313, may provide evidence lated disease. Genetic variants in sodium-dependent Research Institute, Aravind on a causal association of vitamin C with cataract. vitamin C transporter proteins (SVCTs), SLC23A1 Medical Research Foundation, Methods We used data from a randomly selected and SLC23A2 6 are not subject to these concerns Madurai, India 3 population-based study in people aged 60 years and and can provide stronger evidence on the uncon- Cataract Services, Aravind Eye founded association of ascorbate with cataract Hospital, Pondicherry, India above in north and south India. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Differentially Methylated Genes
10/30/2013 Disclosures Key Rheumatoid Arthritis-Associated Pathogenic Pathways Revealed by Integrative Analysis of RA Omics Datasets Consultant: IGNYTA Funding: Rheumatology Research Foundation By John W. Whitaker, Wei Wang and Gary S. Firestein DNA methylation and gene regulation The RA methylation signature in FLS DNA methylation – DNMT1 (maintaining methylation) OA – DNMT3a, 3b (de novo methylation) RA % of CpG methylation: 0% 100% Nakano et al. 2013 ARD AA06 AANAT AARS ABCA6 ABCC12 ABCG1 ABHD8 ABL2 ABR ABRA ACACA ACAN ACAP3 ACCSL ACN9 ACOT7 ACOX2 ACP5 ACP6 ACPP ACSL1 ACSL3 ACSM5 ACVRL1 ADAM10 ADAM32 ADAM33 ADAMTS12 ADAMTS15 ADAMTS19 ADAMTS4 ADAT3 ADCK4 ADCK5 ADCY2 ADCY3 ADCY6 ADORA1 ADPGK ADPRHL1 ADTRP AFAP1 AFAP1L2 AFF3 AFG3L1P AGAP11 AGER AGTR1 AGXT AIF1L AIM2 AIRE AJUBA AK4 AKAP12 AKAP2 AKR1C2 AKR1E2 AKT2 ALAS1 ALDH1L1-AS1 ALDH3A1 ALDH3B1 ALDH8A1 ALDOB ALDOC ALOX12 ALPK3 ALS2CL ALX4 AMBRA1 AMPD2 AMPD3 ANGPT1 ANGPT2 ANGPTL5 ANGPTL6 ANK1 ANKMY2 ANKRD29 ANKRD37 ANKRD53 ANO3 ANO6 ANO7 ANP32C ANXA6 ANXA8L2 AP1G1 AP2A2 AP2M1 AP5B1 APBA2 APC APCDD1 APOBEC3B APOBEC3G APOC1 APOH APOL6 APOLD1 APOM AQP1 AQP10 AQP6 AQP9 ARAP1 ARHGAP24 ARHGAP42 ARHGEF19 ARHGEF25 ARHGEF3 ARHGEF37 ARHGEF7 ARL4C ARL6IP 5 ARL8B ARMC3 ARNTL2 ARPP21 ARRB1 ARSI ASAH2B ASB10 ASB2 ASCL2 ASIC4 ASPH ATF3 ATF7 ATL1 ATL3 ATP10A ATP1A1 ATP1A4 ATP2C1 ATP5A1 ATP5EP2 ATP5L2 ATP6V0CP3 ATP6V1C1 ATP6V1E2 ATXN7L1 ATXN7L2 AVPI1 AXIN2 B3GNT7 B3GNT8 B3GNTL1 BACH1 BAG3 Differential methylated genes in RA FLS BAIAP2L2 BANP BATF BATF2 BBS2 BCAS4 BCAT1 BCL7C BDKRB2 BEGAIN BEST1 BEST3 -

Frontiersin.Org 1 April 2015 | Volume 9 | Article 123 Saunders Et Al
ORIGINAL RESEARCH published: 28 April 2015 doi: 10.3389/fnins.2015.00123 Influx mechanisms in the embryonic and adult rat choroid plexus: a transcriptome study Norman R. Saunders 1*, Katarzyna M. Dziegielewska 1, Kjeld Møllgård 2, Mark D. Habgood 1, Matthew J. Wakefield 3, Helen Lindsay 4, Nathalie Stratzielle 5, Jean-Francois Ghersi-Egea 5 and Shane A. Liddelow 1, 6 1 Department of Pharmacology and Therapeutics, University of Melbourne, Parkville, VIC, Australia, 2 Department of Cellular and Molecular Medicine, University of Copenhagen, Copenhagen, Denmark, 3 Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia, 4 Institute of Molecular Life Sciences, University of Zurich, Zurich, Switzerland, 5 Lyon Neuroscience Research Center, INSERM U1028, Centre National de la Recherche Scientifique UMR5292, Université Lyon 1, Lyon, France, 6 Department of Neurobiology, Stanford University, Stanford, CA, USA The transcriptome of embryonic and adult rat lateral ventricular choroid plexus, using a combination of RNA-Sequencing and microarray data, was analyzed by functional groups of influx transporters, particularly solute carrier (SLC) transporters. RNA-Seq Edited by: Joana A. Palha, was performed at embryonic day (E) 15 and adult with additional data obtained at University of Minho, Portugal intermediate ages from microarray analysis. The largest represented functional group Reviewed by: in the embryo was amino acid transporters (twelve) with expression levels 2–98 times Fernanda Marques, University of Minho, Portugal greater than in the adult. In contrast, in the adult only six amino acid transporters Hanspeter Herzel, were up-regulated compared to the embryo and at more modest enrichment levels Humboldt University, Germany (<5-fold enrichment above E15). -

Modulation of Urate Transport by Drugs
pharmaceutics Review Modulation of Urate Transport by Drugs Péter Tátrai 1, Franciska Erd˝o 2, Gabriella Dörnyei 3 and Péter Krajcsi 1,2,3,* 1 Solvo Biotechnology, Science Park, Building B2, 4-20 Irinyi József utca, H-1117 Budapest, Hungary; [email protected] 2 Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, H-1083 Budapest, Hungary; [email protected] 3 Department of Morphology and Physiology, Faculty of Health Sciences, Semmelweis University, H-1088 Budapest, Hungary; [email protected] * Correspondence: [email protected] Abstract: Background: Serum urate (SU) levels in primates are extraordinarily high among mammals. Urate is a Janus-faced molecule that acts physiologically as a protective antioxidant but provokes inflammation and gout when it precipitates at high concentrations. Transporters play crucial roles in urate disposition, and drugs that interact with urate transporters either by intention or by accident may modulate SU levels. We examined whether in vitro transporter interaction studies may clarify and predict such effects. Methods: Transporter interaction profiles of clinically proven urate-lowering (uricosuric) and hyperuricemic drugs were compiled from the literature, and the predictive value of in vitro-derived cut-offs like Cmax/IC50 on the in vivo outcome (clinically relevant decrease or increase of SU) was assessed. Results: Interaction with the major reabsorptive urate transporter URAT1 appears to be dominant over interactions with secretory transporters in determining the net effect of a drug on SU levels. In vitro inhibition interpreted using the recommended cut-offs is useful at predicting the clinical outcome. Conclusions: In vitro safety assessments regarding urate transport should be done early in drug development to identify candidates at risk of causing major imbalances. -

Ascorbic Acid and the Brain: Rationale for the Use Against Cognitive Decline
Nutrients 2014, 6, 1752-1781; doi:10.3390/nu6041752 OPEN ACCESS nutrients ISSN 2072-6643 www.mdpi.com/journal/nutrients Review Ascorbic Acid and the Brain: Rationale for the Use against Cognitive Decline Fiona E. Harrison 1, Gene L. Bowman 2 and Maria Cristina Polidori 3,* 1 Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN 37232, USA; E-Mail: [email protected] 2 Brain Institute, Department of Neurology, NIA–Aging and Alzheimer’s Disease Center Oregon Health and Science University, Portland, OR 97239, USA; E-Mail: [email protected] 3 Geriatrics Department, University of Cologne Medical Faculty, Cologne 50937, Germany * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +49-221-4788-6704; Fax: +49-221-4788-6710. Received: 25 October 2013; in revised form: 24 March 2014 / Accepted: 10 April 2014 / Published: 24 April 2014 Abstract: This review is focused upon the role of ascorbic acid (AA, vitamin C) in the promotion of healthy brain aging. Particular attention is attributed to the biochemistry and neuronal metabolism interface, transport across tissues, animal models that are useful for this area of research, and the human studies that implicate AA in the continuum between normal cognitive aging and age-related cognitive decline up to Alzheimer’s disease. Vascular risk factors and comorbidity relationships with cognitive decline and AA are discussed to facilitate strategies for advancing AA research in the area of brain health and neurodegeneration. Keywords: ascorbic acid; Vitamin C; brain; cognitive function; alzheimer’s disease; dementia; aging; elderly; endothelial function; blood-brain barrier; SVCT (Sodium-dependent vitamin C transporter) 1.