Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Meta-Analysis and Inding from Seoul Breast Cancer Study (SEBCS)
The Pharmacogenomics Journal https://doi.org/10.1038/s41397-018-0016-6 ARTICLE Associations between genetic polymorphisms of membrane transporter genes and prognosis after chemotherapy: meta-analysis and finding from Seoul Breast Cancer Study (SEBCS) 1 1 1 1 2 3 4 Ji-Eun Kim ● Jaesung Choi ● JooYong Park ● Chulbum Park ● Se Mi Lee ● Seong Eun Park ● Nan Song ● 5 6 4,7 8 1,4,9 10 Seokang Chung ● Hyuna Sung ● Wonshik Han ● Jong Won Lee ● Sue K. Park ● Mi Kyung Kim ● 4,7 9,11 1,4,9,12 1,4,9 Dong-Young Noh ● Keun-Young Yoo ● Daehee Kang ● Ji-Yeob Choi Received: 7 June 2017 / Revised: 13 October 2017 / Accepted: 4 December 2017 © Macmillan Publishers Limited, part of Springer Nature 2018 Abstract Membrane transporters can be major determinants of the pharmacokinetic profiles of anticancer drugs. The associations between genetic variations of ATP-binding cassette (ABC) and solute carrier (SLC) genes and cancer survival were investigated through a meta-analysis and an association study in the Seoul Breast Cancer Study (SEBCS). Including the SEBCS, the meta-analysis was conducted among 38 studies of genetic variations of transporters on various cancer survivors. 1234567890();,: The population of SEBCS consisted of 1 338 breast cancer patients who had been treated with adjuvant chemotherapy. A total of 7 750 SNPs were selected from 453 ABC and/or SLC genes typed by an Affymetrix 6.0 chip. ABCB1 rs1045642 was associated with poor progression-free survival in a meta-analysis (HR = 1.33, 95% CI: 1.07–1.64). ABCB1, SLC8A1, and SLC12A8 were associated with breast cancer survival in SEBCS (Pgene < 0.05). -

New Products JULY 2013
R&D Systems Tools for Cell Biology Research™ New Products JULY 2013 GMP-grade Recombinant Proteins Contents R&D Systems now offers GMP-grade cytokines and growth factors for research and further manu- facturing applications where current Good Manufacturing Practices (GMP) are required. GMP-grade Recombinant Proteins 2 proteins are manufactured in our ISO-certified facility in compliance with relevant guidelines1 and are produced with extensive documentation at every stage of development from cell culture to final fill and Quantikine® ELISA Kits 3 formulation. GMP-grade Recombinant Human IL-6 and Recombinant Human TNF-a have recently been added to our line of GMP-grade proteins. Additional GMP-grade proteins will be available in the next several months. For an up-to-date product listing or additional information, please visit our website at Luminex® Screening Assays 4 www.RnDSystems.com/GMP. Luminex® Performance Assays 4-5 Features New GMP Proteins ✓ Extensive documentation at every stage of development ProtEin SOURCE Catalog # SIZE Polyclonal Antibodies 6-7 ✓ Documentation of lot-to-lot consistency and traceability Human IL-6 E. coli 206-GMP-010 10 µg Monoclonal Antibodies 7-8 of materials used 206-GMP-050 50 µg ✓ Rigorous quality control using stringent analytical 206-GMP-01M 1 mg processes Biotinylated Antibodies 8 Human TNF-a/ E. coli 210-GMP-010 10 µg ✓ Proven formulations to ensure consistent reconstitution TNFSF1A and results 210-GMP-050 50 µg Antibody Controls 8 210-GMP-01M 1 mg ELISpot Kits & Development Modules 8 30000 60 Fluorokine® Flow Cytometry Kits 8 25000 17348 ) 2 50 20000 Fluorochrome-labeled Antibodies 9 40 8673 DuoSet® ELISA and DuoSet IC ELISA 15000 Development Systems 10 30 Peak Intensity Peak 10000 20 Cell-Based ELISA Assay Kits 10 5000 17565 (Mean RFU x10 Viability Cell 8780 10 Parameter Assay Kits 10 0 0 6000 9000 12000 15000 18000 21000 24000 -3 -2 -1 0 1 10 10 10 10 10 Apoptosis Detection 10 Mass Charge Ratio Recombinant Human TNF-α GMP (ng/mL) MALDI-TOF Analysis of GMP-grade Recombinant Human TNF-a. -

Identification of a Novel Nucleobase-Ascorbate Transporter
bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Identification of a novel Nucleobase-Ascorbate 2 Transporter family member in fish and amphibians 3 Diogo Oliveira*,1, André M. Machado*,1, Tiago Cardoso1, Mónica Lopes-Marques1, L. Filipe 4 C. Castro1,2● and Raquel Ruivo1● 5 1CIIMAR – Interdisciplinary Centre of Marine and Environmental Research, U. Porto – University 6 of Porto, Porto, Portugal 7 2Department of Biology, Faculty of Sciences, U. Porto - University of Porto, Portugal 8 *Equal contribution 9 ●Corresponding authors at: CIIMAR, Terminal de Cruzeiros do Porto de Leixões, Av. General 10 Norton de Matos s/n, 4450-208 Matosinhos, Portugal. Tel.: +351 223 401 831 11 12 Running title: Novel uric acid transporter in fish and amphibians 13 14 15 bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 16 Abstract:Nucleobase-Ascorbate Transporter (NAT) family includes ascorbic acid, nucleobases 17 and uric acid transporters: with a broad evolutionary distribution. In vertebrates, four members 18 have been previously recognized, the ascorbate transporters Slc23a1 and Slc3a2, the nucleobase 19 transporter Slc23a4 and an orphan transporter Slc23a3. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Differential Expression of Hydroxyurea Transporters in Normal and Polycythemia Vera Hematopoietic Stem and Progenitor Cell Subpopulations
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2021 Differential expression of hydroxyurea transporters in normal and polycythemia vera hematopoietic stem and progenitor cell subpopulations Tan, Ge ; Meier-Abt, Fabienne Abstract: Polycythemia vera (PV) is a myeloproliferative neoplasm marked by hyperproliferation of the myeloid lineages and the presence of an activating JAK2 mutation. Hydroxyurea (HU) is a standard treat- ment for high-risk patients with PV. Because disease-driving mechanisms are thought to arise in PV stem cells, effective treatments should target primarily the stem cell compartment. We tested for theantipro- liferative effect of patient treatment with HU in fluorescence-activated cell sorting-isolated hematopoietic stem/multipotent progenitor cells (HSC/MPPs) and more committed erythroid progenitors (common myeloid/megakaryocyte-erythrocyte progenitors [CMP/MEPs]) in PV using RNA-sequencing and gene set enrichment analysis. HU treatment led to significant downregulation of gene sets associated with cell proliferation in PV HSCs/MPPs, but not in PV CMP/MEPs. To explore the mechanism underlying this finding, we assessed for expression of solute carrier membrane transporters, which mediate trans- membrane movement of drugs such as HU into target cells. The active HU uptake transporter OCTN1 was upregulated in HSC/MPPs compared with CMP/MEPs of untreated patients with PV, and the HU diffusion facilitator urea transporter B (UTB) was downregulated in HSC/MPPs compared withCM- P/MEPs in all patient and control groups tested. These findings indicate a higher accumulation ofHU within PV HSC/MPPs compared with PV CMP/MEPs and provide an explanation for the differential effects of HU in HSC/MPPs and CMP/MEPs of patients with PV. -
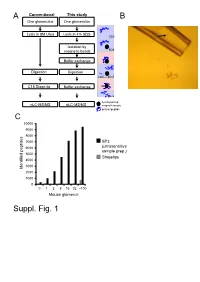
Suppl. Fig. 1 A
A Conventional This study B One glomerulus One glomerulus Lysis in 8M Urea Lysis in 4% SDS SDS Isolation by magnetic beads SDS Buffer exchange Digestion Digestion Try LysC pH>7 C18 Stage tip Buffer exchange pH<4 functionalized nLC-MS/MS nLC-MS/MS magnetic beads protein/peptide C 10000 9000 8000 7000 SP3 6000 (ultrasensitive sample prep.) 5000 Stagetips 4000 3000 Identified peptides 2000 1000 0 0 1 2 8 16 32 ~100 Mouse glomeruli Suppl. Fig. 1 S1. Sample preparation methods. A. Scheme of conventional (C18 Stage Tip) and ultrasensitive proteomic sample preparation methods. B. Manual microdissection and isolation of single glomeruli. The arrow indicates a single isolated glomerulus subjected to proteome analysis. The tip capacity is 10µl. C. Stagetips, a conventional proteomic sample preparation method, is compared with ultrasensitive (SP3) proteome analysis. The indicated amounts of mouse glomeruli were prepared by the respective sample preparation protocols, and peptide numbers after nLC-MS/MS are plotted (all FDR<0.01). A 100 1 Tubule B 80 60 40 Intensity CCD 20 40 0 100 20 S1 TAL 80 0 Tubule proximal tubule Loading... 60 0 Intensity 40 S2 20 -20 control Component 2 (22.5%) S3 0 0 10 20 30 40 50 60 -40 -20 0 20 40 Time (min) Component 2 (47.1%) C D 6000 Mouse proximal tubule 5000 8 4000 3000 7 (iBAQ) Loading... 10 2000 log 6 Number of peptides 1000 0 5 Control mouse mouse mouse human proximal mTAL CCD proximal 0 1500 tubule tubule Rank protein (S1) (S1) R=0.7 E F 8 ACTB Human proximal tubule 8 7 SLC9A3R1 ATP1A1 SLC3A2 SLC25A5 ATP1B1 7 SLC5A2 SLC25A3 CUBN CLTC SLC27A2 SLC5A12 LRP2 6 SLC25A13 SLC25A10 SLC5A1 SLC22A6 6 SLC25A4 Loading.. -

Molecular Profiling of Peripheral Blood Is Associated with Circulating Tumor Cells Content and Poor Survival in Metastatic Castration-Resistant Prostate Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 12 Molecular profiling of peripheral blood is associated with circulating tumor cells content and poor survival in metastatic castration-resistant prostate cancer Mercedes Marín-Aguilera1, Òscar Reig1,2, Juan José Lozano3, Natalia Jiménez1, Susana García-Recio1,4, Nadina Erill5, Lydia Gaba2, Andrea Tagliapietra2, Vanesa Ortega2, Gemma Carrera6, Anna Colomer5, Pedro Gascón4 and Begoña Mellado1,2 1 Translational Genomics Group and Targeted Therapeutics in Solid Tumors Group, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain 2 Medical Oncology Department, Hospital Clínic, Barcelona, Spain 3 Bioinformatics Platform Department, Centro de Investigación Biomédica en Red en el Área temática de Enfermedades Hepáticas y Digestivas (CIBEREHD), Hospital Clínic, Barcelona, Spain 4 Laboratory of Translational Oncology, Fundació Clínic per a la Recerca Biomèdica, Barcelona, Spain 5 Althia, Barcelona, Spain 6 Medical Oncology Department, Hospital Plató, Barcelona, Spain Correspondence to: Begoña Mellado, email: [email protected] Keywords: circulating tumor cells, peripheral blood, microarrays, cell search system Received: January 22, 2015 Accepted: February 14, 2015 Published: March 12, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT The enumeration of circulating -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Supplementary Table 1. All Differentially Expressed Genes from Microarray Screening Analysis
Supplementary Table 1. All differentially expressed genes from microarray screening analysis. FCa (Gal-KD Gene Symbol Gene Name p-Value versus Vector) Up-regulated genes HAPLN1 hyaluronan and proteoglycan link protein 1 10.49 0.0027085 THBS1 thrombospondin 1 9.20 0.0022192 ODC1 ornithine decarboxylase 1 6.38 0.0055776 TGM2 transglutaminase 2 4.76 0.0015627 IL7R interleukin 7 receptor 4.75 0.0017245 SERINC2 serine incorporator 2 4.51 0.0014919 ITM2C integral membrane protein 2C 4.32 0.0044644 SERPINB7 serpin peptidase inhibitor, clade B, member 7 4.18 0.0081136 tumor necrosis factor receptor superfamily, member TNFRSF10D 4.01 0.0085561 10d TAGLN transgelin 3.90 0.0099963 LRRN4 leucine rich repeat neuronal 4 3.82 0.0046513 TGFB2 transforming growth factor beta 2 3.51 0.0035017 CPA4 carboxypeptidase A4 3.43 0.0008452 EPB41L3 erythrocyte membrane protein band 4.1-like 3 3.34 0.0025309 NRG1 neuregulin 1 3.28 0.0079724 F3 coagulation factor III (thromboplastin, tissue factor) 3.27 0.0038968 POLR3G polymerase III polypeptide G 3.26 0.0070675 SEMA7A semaphorin 7A 3.20 0.0087335 NT5E 5-nucleotidase 3.17 0.0036353 CAMK2N1 calmodulin-dependent protein kinase II inhibitor 1 3.07 0.0090141 TIMP3 TIMP metallopeptidase inhibitor 3 3.03 0.0047953 SERPINE1 serpin peptidase inhibitor, clade E 2.97 0.0053652 MALL mal, T-cell differentiation protein-like 2.88 0.0078205 DDAH1 dimethylarginine dimethylaminohydrolase 1 2.86 0.0002895 WDR3 WD repeat domain 3 2.85 0.0058842 WNT5A Wnt Family Member 5A 2.81 0.0043796 GPR1 G protein-coupled receptor 1 2.81 0.0021313 -

Prestinłs Anion Transport and Voltage-Sensing Capabilities Are
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biophysical Journal Volume 96 April 2009 3179–3186 3179 Prestin’s Anion Transport and Voltage-Sensing Capabilities Are Independent Jun-Ping Bai,† Alexei Surguchev,†‡ Simone Montoya,†‡ Peter S. Aronson,§{ Joseph Santos-Sacchi,‡{k and Dhasakumar Navaratnam†k* †Department of Neurology, ‡Division of Otolaryngology, Department of Surgery, §Department of Internal Medicine, {Department of Cellular and Molecular Physiology, and kDepartment of Neurobiology, Yale University School of Medicine, New Haven, Connecticut ABSTRACT The integral membrane protein prestin, a member of the SLC26 anion transporter family, is responsible for the voltage-driven electromotility of mammalian outer hair cells. It was argued that the evolution of prestin’s motor function required a loss of the protein’s transport capabilities. Instead, it was proposed that prestin manages only an abortive hemicycle that results in the trapped anion acting as a voltage sensor, to generate the motor’s signature gating charge movement or nonlinear capaci- tance. We demonstrate, using classical radioactive anion ([14C]formate and [14C]oxalate) uptake studies, that in contrast to previous observations, prestin is able to transport anions. The prestin-dependent uptake of both these anions was twofold that of cells transfected with vector alone, and comparable to SLC26a6, prestin’s closest phylogenetic relative. Furthermore, we identify a potential chloride-binding site in which the mutations of two residues (P328A and L326A) preserve nonlinear capacitance, yet negate anion transport. Finally, we distinguish 12 charged residues out of 22, residing within prestin’s transmembrane regions, that contribute to unitary charge movement, i.e., voltage sensing. -

Taqman® Openarray® Pharmacogenomics (Pgx) Panel
PRODUCT OVERVIEW TaqMan® OpenArray® Pharmacogenomics (PGx) Panel TaqMan® OpenArray® Pharmacogenomics (PGx) Panel Genotyping analysis of drug metabolism enzymes and associated transport proteins The TaqMan® OpenArray® Trusted TaqMan® performance samples, studies, and labs. Assays Pharmacogenomics (PGx) Panel is Each TaqMan® DME Genotyping were selected for optimal relevance a powerful tool to help researchers Assay contains two allele-specific to current pharmacogenomics study human genetic variation probes and a primer pair to detect studies and organized for a simplified in relation to drug action and its the specific SNP target. Both the workflow. potential application to medical probes and primers uniquely align ® ® treatment. within the genome, enabling the The TaqMan OpenArray PGx TaqMan® genotyping technology to Panel provides valuable data for ® ® The TaqMan OpenArray PGx provide superior specificity. It is this the study of drug interactions in Panel was developed for quick and specificity that allows these assays several research areas. Targeted easy screening of known high- to detect targets residing in highly genes relate to areas of study such value target genes associated with homologous gene families that may as cardiovascular (CYP2D6, CYP2C19, drug metabolism enzymes and include pseudogenes. TaqMan® Drug NAT1, NAT2), analgesics (CYP2C9, associated transport proteins. Metabolism Enzyme Genotyping CYP2D6), rheumatology (CYP2C9, The panel consists of 158 drug Assays were developed using a high TPMT), neurology (CYP2C19, CYP2D6), metabolism enzyme (DME) assays level of bioinformatics and wet-lab and musculoskeletal (CYP2C19). A derived from the PharmaADME stringency. All assays have passed total of 29 genes are covered across Core Marker Set (Table 1). The performance tests involving 180 158 unique assays.