Conserved Genes from Simon's Simplex Collection in Drosoph Human Gene HGNC Flybaseid Fly Gene A2M 7 Fbgn00
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Identification of a Novel Nucleobase-Ascorbate Transporter
bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Identification of a novel Nucleobase-Ascorbate 2 Transporter family member in fish and amphibians 3 Diogo Oliveira*,1, André M. Machado*,1, Tiago Cardoso1, Mónica Lopes-Marques1, L. Filipe 4 C. Castro1,2● and Raquel Ruivo1● 5 1CIIMAR – Interdisciplinary Centre of Marine and Environmental Research, U. Porto – University 6 of Porto, Porto, Portugal 7 2Department of Biology, Faculty of Sciences, U. Porto - University of Porto, Portugal 8 *Equal contribution 9 ●Corresponding authors at: CIIMAR, Terminal de Cruzeiros do Porto de Leixões, Av. General 10 Norton de Matos s/n, 4450-208 Matosinhos, Portugal. Tel.: +351 223 401 831 11 12 Running title: Novel uric acid transporter in fish and amphibians 13 14 15 bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 16 Abstract:Nucleobase-Ascorbate Transporter (NAT) family includes ascorbic acid, nucleobases 17 and uric acid transporters: with a broad evolutionary distribution. In vertebrates, four members 18 have been previously recognized, the ascorbate transporters Slc23a1 and Slc3a2, the nucleobase 19 transporter Slc23a4 and an orphan transporter Slc23a3. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Differential Expression of Hydroxyurea Transporters in Normal and Polycythemia Vera Hematopoietic Stem and Progenitor Cell Subpopulations
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2021 Differential expression of hydroxyurea transporters in normal and polycythemia vera hematopoietic stem and progenitor cell subpopulations Tan, Ge ; Meier-Abt, Fabienne Abstract: Polycythemia vera (PV) is a myeloproliferative neoplasm marked by hyperproliferation of the myeloid lineages and the presence of an activating JAK2 mutation. Hydroxyurea (HU) is a standard treat- ment for high-risk patients with PV. Because disease-driving mechanisms are thought to arise in PV stem cells, effective treatments should target primarily the stem cell compartment. We tested for theantipro- liferative effect of patient treatment with HU in fluorescence-activated cell sorting-isolated hematopoietic stem/multipotent progenitor cells (HSC/MPPs) and more committed erythroid progenitors (common myeloid/megakaryocyte-erythrocyte progenitors [CMP/MEPs]) in PV using RNA-sequencing and gene set enrichment analysis. HU treatment led to significant downregulation of gene sets associated with cell proliferation in PV HSCs/MPPs, but not in PV CMP/MEPs. To explore the mechanism underlying this finding, we assessed for expression of solute carrier membrane transporters, which mediate trans- membrane movement of drugs such as HU into target cells. The active HU uptake transporter OCTN1 was upregulated in HSC/MPPs compared with CMP/MEPs of untreated patients with PV, and the HU diffusion facilitator urea transporter B (UTB) was downregulated in HSC/MPPs compared withCM- P/MEPs in all patient and control groups tested. These findings indicate a higher accumulation ofHU within PV HSC/MPPs compared with PV CMP/MEPs and provide an explanation for the differential effects of HU in HSC/MPPs and CMP/MEPs of patients with PV. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -
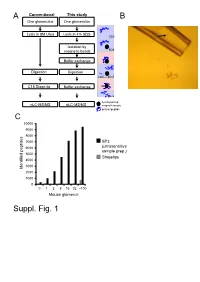
Suppl. Fig. 1 A
A Conventional This study B One glomerulus One glomerulus Lysis in 8M Urea Lysis in 4% SDS SDS Isolation by magnetic beads SDS Buffer exchange Digestion Digestion Try LysC pH>7 C18 Stage tip Buffer exchange pH<4 functionalized nLC-MS/MS nLC-MS/MS magnetic beads protein/peptide C 10000 9000 8000 7000 SP3 6000 (ultrasensitive sample prep.) 5000 Stagetips 4000 3000 Identified peptides 2000 1000 0 0 1 2 8 16 32 ~100 Mouse glomeruli Suppl. Fig. 1 S1. Sample preparation methods. A. Scheme of conventional (C18 Stage Tip) and ultrasensitive proteomic sample preparation methods. B. Manual microdissection and isolation of single glomeruli. The arrow indicates a single isolated glomerulus subjected to proteome analysis. The tip capacity is 10µl. C. Stagetips, a conventional proteomic sample preparation method, is compared with ultrasensitive (SP3) proteome analysis. The indicated amounts of mouse glomeruli were prepared by the respective sample preparation protocols, and peptide numbers after nLC-MS/MS are plotted (all FDR<0.01). A 100 1 Tubule B 80 60 40 Intensity CCD 20 40 0 100 20 S1 TAL 80 0 Tubule proximal tubule Loading... 60 0 Intensity 40 S2 20 -20 control Component 2 (22.5%) S3 0 0 10 20 30 40 50 60 -40 -20 0 20 40 Time (min) Component 2 (47.1%) C D 6000 Mouse proximal tubule 5000 8 4000 3000 7 (iBAQ) Loading... 10 2000 log 6 Number of peptides 1000 0 5 Control mouse mouse mouse human proximal mTAL CCD proximal 0 1500 tubule tubule Rank protein (S1) (S1) R=0.7 E F 8 ACTB Human proximal tubule 8 7 SLC9A3R1 ATP1A1 SLC3A2 SLC25A5 ATP1B1 7 SLC5A2 SLC25A3 CUBN CLTC SLC27A2 SLC5A12 LRP2 6 SLC25A13 SLC25A10 SLC5A1 SLC22A6 6 SLC25A4 Loading.. -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Role of CREB/CRTC1-Regulated Gene Transcription During Hippocampal-Dependent Memory in Alzheimer’S Disease Mouse Models
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Institut de Neurociències Universitat Autònoma de Barcelona Departament de Bioquímica i Biologia Molecular Unitat de Bioquímica, Facultat de Medicina Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Arnaldo J. Parra Damas TESIS DOCTORAL Bellaterra, 2015 Institut de Neurociències Departament de Bioquímica i Biologia Molecular Universitat Autònoma de Barcelona Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Papel de la transcripción génica regulada por CRTC1/CREB durante memoria dependiente de hipocampo en modelos murinos de la enfermedad de Alzheimer Memoria de tesis doctoral presentada por Arnaldo J. Parra Damas para optar al grado de Doctor en Neurociencias por la Universitat Autonòma de Barcelona. Trabajo realizado en la Unidad de Bioquímica y Biología Molecular de la Facultad de Medicina del Departamento de Bioquímica y Biología Molecular de la Universitat Autònoma de Barcelona, y en el Instituto de Neurociencias de la Universitat Autònoma de Barcelona, bajo la dirección del Doctor Carlos Saura Antolín. -

9-Azido Analogs of Three Sialic Acid Forms for Metabolic Remodeling Of
Supporting Information 9-Azido Analogs of Three Sialic Acid Forms for Metabolic Remodeling of Cell-Surface Sialoglycans Bo Cheng,†,‡ Lu Dong,†,§ Yuntao Zhu,†,‡ Rongbing Huang,†,‡ Yuting Sun,†,‖ Qiancheng You,†,‡ Qitao Song,†,§ James C. Paton, ∇ Adrienne W. Paton,∇ and Xing Chen*,†,‡,§,⊥,# †College of Chemistry and Molecular Engineering, ‡Beijing National Laboratory for Molecular Sciences, §Peking−Tsinghua Center for Life Sciences,‖Academy for Advanced Interdisciplinary Studies, ⊥Synthetic and Functional Biomolecules Center, and #Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, Peking University, Beijing 100871, China ∇Research Centre for Infectious Diseases, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide SA 5005, Australia Page S1 Table of Contents: Scheme S1.……………………………………………………….........……………. S3 Figure S1……………………………………………………..………..……………. S3 Figure S2……………………………………………………..………..…………… S4 Figure S3……………………………………………………..………..…………… S4 Figure S4……………………………………………………..………..…………… S5 Figure S5……………………………………………………..………..…………… S6 Figure S6……………………………………………………..………..…………….S7 Figure S7……………………………………………………..………..…………….S8 Figure S8……………………………………………………..………..…………….S9 Experimental Procedures……………………………….…........…………....S10-S27 Table S1………………………………………………..………..…………….S28-S48 Supporting Reference……………………………………………….......………...S49 Page S2 Scheme S1. Synthesis of 9AzNeu5Gc Figure S1: a, b, c, d) Representative scatter plots (FSC vs. SSC) and histograms of flow cytometry analysis -

Genomic Dissection of 43 Serum Urate-Associated Loci Provides
bioRxiv preprint doi: https://doi.org/10.1101/743864; this version posted August 22, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Genomic dissection of 43 serum urate-associated loci provides 2 multiple insights into molecular mechanisms of urate control. 3 4 James Boocock1,2¶, Megan Leask1¶, Yukinori Okada3,4, Asian Genetic Epidemiology 5 Network (AGEN) Consortium, Hirotaka Matsuo5, Yusuke Kawamura5, Yongyong 6 Shi6, Changgui Li7, David B Mount8,9, Asim K Mandal8, Weiqing Wang10, Murray 7 Cadzow1, Anna L Gosling1, Tanya J Major1, Julia A Horsfield11, Hyon K Choi12, 8 Tayaza Fadason13, Justin O’Sullivan13, Eli A Stahl10&, Tony R Merriman1*& 9 10 1 Department of Biochemistry, Biomedical Sciences, University of Otago, Dunedin, 11 New Zealand 12 2 Department of Human Genetics, David Geffen School of Medicine at UCLA, Los 13 Angeles, CA, USA 14 3 Department of Statistical Genetics, Osaka University Graduate School of Medicine, 15 Osaka, Japan 16 4 Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI- 17 IFReC), Osaka University, Suita, Japan 18 5 Department of Integrative Physiology and Bio-Nano Medicine, National Defense 19 Medical College, Tokorozawa, Saitama, Japan 20 6 Bio-X Institutes, Key Laboratory for the Genetics of Developmental and 21 Neuropsychiaric Disorders (Ministry of Education), Shanghai Jiao Tong University, 22 Shanghai, People's Republic of China 23 7 The Department of Endocrinology -

CREB-Regulated Transcription Coactivator 3 (CRTC3) Polymorphism Associated with Type 2 Diabetes and Hyperlipidemia in the Taiwanese Population
Tzu Chi Medical Journal 26 (2014) 114e118 Contents lists available at ScienceDirect Tzu Chi Medical Journal journal homepage: www.tzuchimedjnl.com Original Article CREB-regulated transcription coactivator 3 (CRTC3) polymorphism associated with type 2 diabetes and hyperlipidemia in the Taiwanese population y y Kuei-Fang Lee a, b, , Cheng-Chia Lin c, , Tsung-Cheng Hsieh a, Chun-Te Wu c, * Lawrence Shih-Hsin Wu a, a Institute of Medical Sciences, Tzu Chi University, Hualien, Taiwan b Laboratory for Cytogenetics, Center for Genetic Counseling, Buddhist Tzu Chi General Hospital, Hualien, Taiwan c Department of Urological Surgery, Chang Gung Memorial Hospital, Keelung, Taiwan article info abstract Article history: Objective: Type 2 diabetes mellitus (T2D) is a pathologically and genetically heterogeneous disease Received 24 April 2014 influenced by genetic and environmental factors. This study aims to investigate the association between Received in revised form T2D and polymorphism(s) in CREB-regulated transcription coactivator 3 (CRTC3) in Asian Taiwan. 27 May 2014 Materials and methods: In this study, 417 participants with T2D and 197 without T2D were recruited. Accepted 9 July 2014 Anthropometrics, the metabolic profile, blood pressure, fasting plasma glucose, glycosylated hemoglobin (HbA1c), serum triglycerides, serum total cholesterol, low-density lipoprotein, high-density lipoprotein Keywords: (HDL), and C-peptide were analyzed. TaqMan genotyping was used to identify individual genotypes, and CRTC3 Hyperlipidemia the association of CRTC3 polymorphism with clinical and biochemical parameters was assessed. SNP Results: Single nucleotide polymorphism (SNP) rs8033595 showed an association with diabetes Type 2 diabetes (p ¼ 0.031) and hyperlipidemia (p ¼ 0.002). Odds ratio analysis showed that A carriers (AA or AG) had a protective effect against developing T2D and hyperlipidemia.