1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Genetic Screening Identifies a Component of the SWI/SNF Complex, Arid1b As a Senescence Regulator
A genetic screening identifies a component of the SWI/SNF complex, Arid1b as a senescence regulator Sadaf Khan A thesis submitted to Imperial College London for the degree of Doctor in Philosophy MRC Clinical Sciences Centre Imperial College London, School of Medicine July 2013 Statement of originality All experiments included in this thesis were performed by myself unless otherwise stated. Copyright Declaration The copyright of this thesis rests with the author and is made available under a Creative Commons Attribution Non-Commercial No Derivatives license. Researchers are free to copy, distribute or transmit the thesis on the condition that they attribute it, that they do not use it for commercial purposes and that they do not alter, transform or build upon it. For any reuse or redistribution, researchers must make clear to others the license terms of this work. 2 Abstract Senescence is an important tumour suppressor mechanism, which prevents the proliferation of stressed or damaged cells. The use of RNA interference to identify genes with a role in senescence is an important tool in the discovery of novel cancer genes. In this work, a protocol was established for conducting bypass of senescence screenings, using shRNA libraries together with next-generation sequencing. Using this approach, the SWI/SNF subunit Arid1b was identified as a regulator of cellular lifespan in MEFs. SWI/SNF is a large multi-subunit complex that remodels chromatin. Mutations in SWI/SNF proteins are frequently associated with cancer, suggesting that SWI/SNF components are tumour suppressors. Here the role of ARID1B during senescence was investigated. Depletion of ARID1B extends the proliferative capacity of primary mouse and human fibroblasts. -

Aberrant Methylation Underlies Insulin Gene Expression in Human Insulinoma
ARTICLE https://doi.org/10.1038/s41467-020-18839-1 OPEN Aberrant methylation underlies insulin gene expression in human insulinoma Esra Karakose1,6, Huan Wang 2,6, William Inabnet1, Rajesh V. Thakker 3, Steven Libutti4, Gustavo Fernandez-Ranvier 1, Hyunsuk Suh1, Mark Stevenson 3, Yayoi Kinoshita1, Michael Donovan1, Yevgeniy Antipin1,2, Yan Li5, Xiaoxiao Liu 5, Fulai Jin 5, Peng Wang 1, Andrew Uzilov 1,2, ✉ Carmen Argmann 1, Eric E. Schadt 1,2, Andrew F. Stewart 1,7 , Donald K. Scott 1,7 & Luca Lambertini 1,6 1234567890():,; Human insulinomas are rare, benign, slowly proliferating, insulin-producing beta cell tumors that provide a molecular “recipe” or “roadmap” for pathways that control human beta cell regeneration. An earlier study revealed abnormal methylation in the imprinted p15.5-p15.4 region of chromosome 11, known to be abnormally methylated in another disorder of expanded beta cell mass and function: the focal variant of congenital hyperinsulinism. Here, we compare deep DNA methylome sequencing on 19 human insulinomas, and five sets of normal beta cells. We find a remarkably consistent, abnormal methylation pattern in insu- linomas. The findings suggest that abnormal insulin (INS) promoter methylation and altered transcription factor expression create alternative drivers of INS expression, replacing cano- nical PDX1-driven beta cell specification with a pathological, looping, distal enhancer-based form of transcriptional regulation. Finally, NFaT transcription factors, rather than the cano- nical PDX1 enhancer complex, are predicted to drive INS transactivation. 1 From the Diabetes Obesity and Metabolism Institute, The Department of Surgery, The Department of Pathology, The Department of Genetics and Genomics Sciences and The Institute for Genomics and Multiscale Biology, The Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -
Nucleoporin 107, 62 and 153 Mediate Kcnq1ot1 Imprinted Domain Regulation in Extraembryonic Endoderm Stem Cells
ARTICLE DOI: 10.1038/s41467-018-05208-2 OPEN Nucleoporin 107, 62 and 153 mediate Kcnq1ot1 imprinted domain regulation in extraembryonic endoderm stem cells Saqib S. Sachani 1,2,3,4, Lauren S. Landschoot1,2, Liyue Zhang1,2, Carlee R. White1,2, William A. MacDonald3,4, Michael C. Golding 5 & Mellissa R.W. Mann 3,4 1234567890():,; Genomic imprinting is a phenomenon that restricts transcription to predominantly one par- ental allele. How this transcriptional duality is regulated is poorly understood. Here we perform an RNA interference screen for epigenetic factors involved in paternal allelic silen- cing at the Kcnq1ot1 imprinted domain in mouse extraembryonic endoderm stem cells. Multiple factors are identified, including nucleoporin 107 (NUP107). To determine NUP107’s role and specificity in Kcnq1ot1 imprinted domain regulation, we deplete Nup107, as well as Nup62, Nup98/96 and Nup153. Nup107, Nup62 and Nup153, but not Nup98/96 depletion, reduce Kcnq1ot1 noncoding RNA volume, displace the Kcnq1ot1 domain from the nuclear periphery, reactivate a subset of normally silent paternal alleles in the domain, alter histone modifications with concomitant changes in KMT2A, EZH2 and EHMT2 occupancy, as well as reduce cohesin interactions at the Kcnq1ot1 imprinting control region. Our results establish an important role for specific nucleoporins in mediating Kcnq1ot1 imprinted domain regulation. 1 Departments of Obstetrics & Gynaecology, and Biochemistry, Western University, Schulich School of Medicine and Dentistry, London, ON N6A 5W9, Canada. 2 Children’s Health Research Institute, London, ON N6C 2V5, Canada. 3 Departments of Obstetrics, Gynecology and Reproductive Sciences, University of Pittsburgh School of Medicine, Pittsburgh, PA 15213, USA. 4 Magee-Womens Research Institute, Pittsburgh, PA 15213, USA. -

Mir-338-3P Functions As a Tumor Suppressor in Gastric Cancer by Targeting PTP1B
Sun et al. Cell Death and Disease DOI 10.1038/s41419-018-0611-0 Cell Death & Disease ARTICLE Open Access miR-338-3p functions as a tumor suppressor in gastric cancer by targeting PTP1B Feng Sun1, Mengchao Yu2,JingYu2, Zhijian Liu1,XinyanZhou2,YanqingLiu2, Xiaolong Ge3,HaidongGao2, Mei Li4, Xiaohong Jiang2,SongLiu1,XiChen2 and Wenxian Guan 1 Abstract Gastric cancer (GC) is one of the most common malignant tumors and peritoneal metastasis is the primary cause for advanced GC’s mortality. Protein-tyrosine phosphatase 1B (PTP1B) functions as an oncogene and involves in carcinogenesis and cancer dissemination. However, the function and regulation of PTP1B in GC remain poorly understood. In this study, we found that PTP1B was upregulated in GC tissues and overexpression of PTP1B in vitro promoted cell migration and prevented apoptosis. Then, we predicted that PTP1B was a target of miR-338-3p and we revealed an inverse correlation between miR-338-3p levels and PTP1B protein levels in GC tissues. Next, we verified that PTP1B was inhibited by miR-338-3p via direct targeting to its 3′-untranslated regions. Moreover, overexpression of miR-338-3p in vitro attenuated GC cell migration and promoted apoptosis, and these effects could be partially reversed by reintroduction of PTP1B. Finally, we established an orthotopic xenograft model and a peritoneal dissemination model of GC to demonstrate that miR-338-3p restrained tumor growth and dissemination in vivo by targeting PTP1B. Taken together, our results highlight that PTP1B is an oncogene and is negatively regulated by miR- 1234567890():,; 1234567890():,; 338-3p in GC, which may provide new insights into novel molecular therapeutic targets for GC. -

Table S1. List of Proteins in the BAHD1 Interactome
Table S1. List of proteins in the BAHD1 interactome BAHD1 nuclear partners found in this work yeast two-hybrid screen Name Description Function Reference (a) Chromatin adapters HP1α (CBX5) chromobox homolog 5 (HP1 alpha) Binds histone H3 methylated on lysine 9 and chromatin-associated proteins (20-23) HP1β (CBX1) chromobox homolog 1 (HP1 beta) Binds histone H3 methylated on lysine 9 and chromatin-associated proteins HP1γ (CBX3) chromobox homolog 3 (HP1 gamma) Binds histone H3 methylated on lysine 9 and chromatin-associated proteins MBD1 methyl-CpG binding domain protein 1 Binds methylated CpG dinucleotide and chromatin-associated proteins (22, 24-26) Chromatin modification enzymes CHD1 chromodomain helicase DNA binding protein 1 ATP-dependent chromatin remodeling activity (27-28) HDAC5 histone deacetylase 5 Histone deacetylase activity (23,29,30) SETDB1 (ESET;KMT1E) SET domain, bifurcated 1 Histone-lysine N-methyltransferase activity (31-34) Transcription factors GTF3C2 general transcription factor IIIC, polypeptide 2, beta 110kDa Required for RNA polymerase III-mediated transcription HEYL (Hey3) hairy/enhancer-of-split related with YRPW motif-like DNA-binding transcription factor with basic helix-loop-helix domain (35) KLF10 (TIEG1) Kruppel-like factor 10 DNA-binding transcription factor with C2H2 zinc finger domain (36) NR2F1 (COUP-TFI) nuclear receptor subfamily 2, group F, member 1 DNA-binding transcription factor with C4 type zinc finger domain (ligand-regulated) (36) PEG3 paternally expressed 3 DNA-binding transcription factor with -
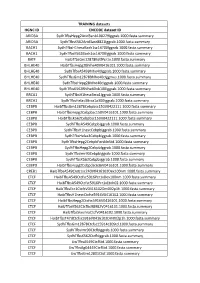
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

ZNF652, a Novel Zinc Finger Protein, Interacts with the Putative Breast Tumor Suppressor CBFA2T3 to Repress Transcription
ZNF652, A Novel Zinc Finger Protein, Interacts with the Putative Breast Tumor Suppressor CBFA2T3 to Repress Transcription Raman Kumar,1 Jantina Manning,1 Hayley E. Spendlove,3 Gabriel Kremmidiotis,4 Ross McKirdy,1 Jaclyn Lee,1 David N. Millband,1 Kelly M. Cheney,1 Martha R. Stampfer,5 Prem P. Dwivedi,2 Howard A. Morris,2 and David F. Callen1 1Breast Cancer Genetics Group, Dame Roma Mitchell Cancer Research Laboratories, Department of Medicine, University of Adelaide and Hanson Institute; 2Endocrine Bone Laboratory, Hanson Institute, Adelaide, South Australia, Australia; 3Department of Laboratory Genetics, Women’s and Children’s Hospital, North Adelaide, South Australia, Australia; 4Bionomics, Ltd., Thebarton, South Australia, Australia; and 5Lawrence Berkeley National Laboratory, Berkeley, California Abstract gene effector zinc finger proteins may specifically The transcriptional repressor CBFA2T3is a putative interact with one or more of the ETO proteins to generate breast tumor suppressor. To define the role of CBFA2T3, a defined range of transcriptional repressor complexes. we used a segment of this protein as bait in a yeast (Mol Cancer Res 2006;4(9):655–65) two-hybrid screen and identified a novel uncharacterized protein, ZNF652. In general, primary tumors and cancer Introduction cell lines showed lower expression of ZNF652 than Tumor growth, characterized by unchecked cell division, normal tissues. Together with the location of this gene results from both the overexpression of growth-promoting on the long arm of chromosome 17q, a region of frequent oncogenes and the reduced expression of growth-inhibiting loss of heterozygosity in cancer, these results suggest tumor suppressor genes. These genes often encode proteins that In silico a possible role of ZNF652 in tumorigenesis. -

Genetic Variability in the Italian Heavy Draught Horse from Pedigree Data and Genomic Information
Supplementary material for manuscript: Genetic variability in the Italian Heavy Draught Horse from pedigree data and genomic information. Enrico Mancin†, Michela Ablondi†, Roberto Mantovani*, Giuseppe Pigozzi, Alberto Sabbioni and Cristina Sartori ** Correspondence: [email protected] † These two Authors equally contributed to the work Supplementary Figure S1. Mares and foal of Italian Heavy Draught Horse (IHDH; courtesy of Cinzia Stoppa) Supplementary Figure S2. Number of Equivalent Generations (EqGen; above) and pedigree completeness (PC; below) over years in Italian Heavy Draught Horse population. Supplementary Table S1. Descriptive statistics of homozygosity (observed: Ho_obs; expected: Ho_exp; total: Ho_tot) in 267 genotyped individuals of Italian Heavy Draught Horse based on the number of homozygous genotypes. Parameter Mean SD Min Max Ho_obs 35,630.3 500.7 34,291 38,013 Ho_exp 35,707.8 64.0 35,010 35,740 Ho_tot 50,674.5 93.8 49,638 50,714 1 Definitions of the methods for inbreeding are in the text. Supplementary Figure S3. Values of BIC obtained by analyzing values of K from 1 to 10, corresponding on the same amount of clusters defining the proportion of ancestry in the 267 genotyped individuals. Supplementary Table S2. Estimation of genomic effective population size (Ne) traced back to 18 generations ago (Gen. ago). The linkage disequilibrium estimation, adjusted for sampling bias was also included (LD_r2), as well as the relative standard deviation (SD(LD_r2)). Gen. ago Ne LD_r2 SD(LD_r2) 1 100 0.009 0.014 2 108 0.011 0.018 3 118 0.015 0.024 4 126 0.017 0.028 5 134 0.019 0.031 6 143 0.021 0.034 7 156 0.023 0.038 9 173 0.026 0.041 11 189 0.029 0.046 14 213 0.032 0.052 18 241 0.036 0.058 Supplementary Table S3. -

BAP1: Case Report and Insight Into a Novel Tumor Suppressor Kanad Ghosh1, Badri Modi2, William D
Ghosh et al. BMC Dermatology (2017) 17:13 DOI 10.1186/s12895-017-0065-6 CASEREPORT Open Access BAP1: case report and insight into a novel tumor suppressor Kanad Ghosh1, Badri Modi2, William D. James2 and Brian C. Capell1,2,3* Abstract Background: BRCA1-Associated-Protein 1 (BAP1) is a dynamic tumor suppressor which, when mutated, has been associated with an increased risk of uveal melanoma, cutaneous melanoma, mesothelioma, and several other cancers. Germline BAP1 mutations have been extensively studied, where they have been found to cause hereditary cancer susceptibility. However, their sporadic counterparts, tumors that display a loss of BAP1 expression due to somatically arising mutations in the BAP1 gene, remain a poorly described entity. Case presentation: Here we present the case of a 49-year-old female who presented with an asymptomatic dome-shaped pink papule on the dorsal foot which was found on biopsy to be deficient in the BAP1 tumor suppressor. While the patient’s family history did not suggest the presence of a familial cancer syndrome, germline genetic testing was performed and was negative. The patient underwent surgical excision of this sporadically appearing “BAPoma” by Mohs surgery. Conclusions: Given the relatively banal clinical appearance of these dome-shaped neoplasms, sporadic BAPomas may often be overlooked by clinicians and dermatologists. In addition to providing a representative case, here we also provide a synopsis of the current understanding of these neoplasms, both in terms of the histopathological features, as well as the molecular mechanisms underlying BAP1 function and its ability to prevent tumorigenesis. Keywords: BAP1, Tumor suppression, Familial cancer syndrome Background More recently, sporadic somatic BAP1 mutations have Within the last decade, the BRCA1-Associated-Protein 1 been shown to occur in the setting of both mesothelioma (BAP1) has been increasingly appreciated for its tumor sup- and uveal melanoma [3]. -

ARID1B Is a Specific Vulnerability in ARID1A-Mutant Cancers The
ARID1B is a specific vulnerability in ARID1A-mutant cancers The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Helming, K. C., X. Wang, B. G. Wilson, F. Vazquez, J. R. Haswell, H. E. Manchester, Y. Kim, et al. 2014. “ARID1B is a specific vulnerability in ARID1A-mutant cancers.” Nature medicine 20 (3): 251-254. doi:10.1038/nm.3480. http://dx.doi.org/10.1038/nm.3480. Published Version doi:10.1038/nm.3480 Accessed February 16, 2015 10:04:32 PM EST Citable Link http://nrs.harvard.edu/urn-3:HUL.InstRepos:12987227 Terms of Use This article was downloaded from Harvard University's DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http://nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA (Article begins on next page) NIH Public Access Author Manuscript Nat Med. Author manuscript; available in PMC 2014 September 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Nat Med. 2014 March ; 20(3): 251–254. doi:10.1038/nm.3480. ARID1B is a specific vulnerability in ARID1A-mutant cancers Katherine C. Helming1,2,3,4,*, Xiaofeng Wang1,2,3,*, Boris G. Wilson1,2,3, Francisca Vazquez5, Jeffrey R. Haswell1,2,3, Haley E. Manchester1,2,3, Youngha Kim1,2,3, Gregory V. Kryukov5, Mahmoud Ghandi5, Andrew J. Aguirre5,6,7, Zainab Jagani8, Zhong Wang9, Levi A. Garraway6, William C. Hahn6,7, and Charles W. -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7