Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Expression Polarization
Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression This information is current as of September 27, 2021. Fernando O. Martinez, Siamon Gordon, Massimo Locati and Alberto Mantovani J Immunol 2006; 177:7303-7311; ; doi: 10.4049/jimmunol.177.10.7303 http://www.jimmunol.org/content/177/10/7303 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2006/11/03/177.10.7303.DC1 Material http://www.jimmunol.org/ References This article cites 61 articles, 22 of which you can access for free at: http://www.jimmunol.org/content/177/10/7303.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 27, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression1 Fernando O. -
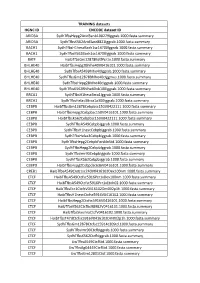
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

Multiple Activities of Arl1 Gtpase in the Trans-Golgi Network Chia-Jung Yu1,2 and Fang-Jen S
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1691-1699 doi:10.1242/jcs.201319 COMMENTARY Multiple activities of Arl1 GTPase in the trans-Golgi network Chia-Jung Yu1,2 and Fang-Jen S. Lee3,4,* ABSTRACT typical features of an Arf-family GTPase, including an amphipathic ADP-ribosylation factors (Arfs) and ADP-ribosylation factor-like N-terminal helix and a consensus site for N-myristoylation (Lu et al., proteins (Arls) are highly conserved small GTPases that function 2001; Price et al., 2005). In yeast, recruitment of Arl1 to the Golgi as main regulators of vesicular trafficking and cytoskeletal complex requires a second Arf-like GTPase, Arl3 (Behnia et al., reorganization. Arl1, the first identified member of the large Arl family, 2004; Setty et al., 2003). Yeast Arl3 lacks a myristoylation site and is an important regulator of Golgi complex structure and function in is, instead, N-terminally acetylated; this modification is required for organisms ranging from yeast to mammals. Together with its effectors, its recruitment to the Golgi complex by Sys1. In mammalian cells, Arl1 has been shown to be involved in several cellular processes, ADP-ribosylation-factor-related protein 1 (Arfrp1), a mammalian including endosomal trans-Golgi network and secretory trafficking, lipid ortholog of yeast Arl3, plays a pivotal role in the recruitment of Arl1 droplet and salivary granule formation, innate immunity and neuronal to the trans-Golgi network (TGN) (Behnia et al., 2004; Panic et al., development, stress tolerance, as well as the response of the unfolded 2003b; Setty et al., 2003; Zahn et al., 2006). -

SPATA33 Localizes Calcineurin to the Mitochondria and Regulates Sperm Motility in Mice
SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice Haruhiko Miyataa, Seiya Ouraa,b, Akane Morohoshia,c, Keisuke Shimadaa, Daisuke Mashikoa,1, Yuki Oyamaa,b, Yuki Kanedaa,b, Takafumi Matsumuraa,2, Ferheen Abbasia,3, and Masahito Ikawaa,b,c,d,4 aResearch Institute for Microbial Diseases, Osaka University, Osaka 5650871, Japan; bGraduate School of Pharmaceutical Sciences, Osaka University, Osaka 5650871, Japan; cGraduate School of Medicine, Osaka University, Osaka 5650871, Japan; and dThe Institute of Medical Science, The University of Tokyo, Tokyo 1088639, Japan Edited by Mariana F. Wolfner, Cornell University, Ithaca, NY, and approved July 27, 2021 (received for review April 8, 2021) Calcineurin is a calcium-dependent phosphatase that plays roles in calcineurin can be a target for reversible and rapidly acting male a variety of biological processes including immune responses. In sper- contraceptives (5). However, it is challenging to develop molecules matozoa, there is a testis-enriched calcineurin composed of PPP3CC and that specifically inhibit sperm calcineurin and not somatic calci- PPP3R2 (sperm calcineurin) that is essential for sperm motility and male neurin because of sequence similarities (82% amino acid identity fertility. Because sperm calcineurin has been proposed as a target for between human PPP3CA and PPP3CC and 85% amino acid reversible male contraceptives, identifying proteins that interact with identity between human PPP3R1 and PPP3R2). Therefore, identi- sperm calcineurin widens the choice for developing specific inhibitors. fying proteins that interact with sperm calcineurin widens the choice Here, by screening the calcineurin-interacting PxIxIT consensus motif of inhibitors that target the sperm calcineurin pathway. in silico and analyzing the function of candidate proteins through the The PxIxIT motif is a conserved sequence found in generation of gene-modified mice, we discovered that SPATA33 inter- calcineurin-binding proteins (8, 9). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Mammalian Male Germ Cells Are Fertile Ground for Expression Profiling Of
REPRODUCTIONREVIEW Mammalian male germ cells are fertile ground for expression profiling of sexual reproduction Gunnar Wrobel and Michael Primig Biozentrum and Swiss Institute of Bioinformatics, Klingelbergstrasse 50-70, 4056 Basel, Switzerland Correspondence should be addressed to Michael Primig; Email: [email protected] Abstract Recent large-scale transcriptional profiling experiments of mammalian spermatogenesis using rodent model systems and different types of microarrays have yielded insight into the expression program of male germ cells. These studies revealed that an astonishingly large number of loci are differentially expressed during spermatogenesis. Among them are several hundred transcripts that appear to be specific for meiotic and post-meiotic germ cells. This group includes many genes that were pre- viously implicated in spermatogenesis and/or fertility and others that are as yet poorly characterized. Profiling experiments thus reveal candidates for regulation of spermatogenesis and fertility as well as targets for innovative contraceptives that act on gene products absent in somatic tissues. In this review, consolidated high density oligonucleotide microarray data from rodent total testis and purified germ cell samples are analyzed and their impact on our understanding of the transcriptional program governing male germ cell differentiation is discussed. Reproduction (2005) 129 1–7 Introduction 2002, Sadate-Ngatchou et al. 2003) and the absence of cAMP responsive-element modulator (Crem) and deleted During mammalian male -

The Flagellar Arginine Kinase in Trypanosoma Brucei Is Important for Infection in Tsetse Flies
RESEARCH ARTICLE The Flagellar Arginine Kinase in Trypanosoma brucei Is Important for Infection in Tsetse Flies Cher-Pheng Ooi1¤, Brice Rotureau1, Simonetta Gribaldo2, Christina Georgikou1, Daria Julkowska1, Thierry Blisnick1, Sylvie Perrot1, Ines Subota1, Philippe Bastin1* 1 Trypanosome Cell Biology Unit, INSERM U1201, Institut Pasteur, 25 Rue du Docteur Roux, 75015, Paris, France, 2 Molecular Biology of Gene in Extremophiles Unit, Department of Microbiology, Institut Pasteur, 25 rue du Docteur Roux, 75015, Paris, France ¤ Current address: Department of Life Sciences, Sir Alexander Fleming Building, Imperial College-South Kensington, London, SW7 2AZ, United Kingdom * [email protected] Abstract OPEN ACCESS African trypanosomes are flagellated parasites that cause sleeping sickness. Parasites are Citation: Ooi C-P, Rotureau B, Gribaldo S, Georgikou C, Julkowska D, Blisnick T, et al. (2015) transmitted from one mammalian host to another by the bite of a tsetse fly. Trypanosoma The Flagellar Arginine Kinase in Trypanosoma brucei brucei possesses three different genes for arginine kinase (AK) including one (AK3) that Is Important for Infection in Tsetse Flies. PLoS ONE encodes a protein localised to the flagellum. AK3 is characterised by the presence of a 10(7): e0133676. doi:10.1371/journal.pone.0133676 unique amino-terminal insertion that specifies flagellar targeting. We show here a phyloge- Editor: Frank Voncken, University of Hull, UNITED netic analysis revealing that flagellar AK arose in two independent duplication events in KINGDOM T. brucei and T. congolense, the two species of African trypanosomes that infect the tsetse Received: April 11, 2015 midgut. In T. brucei, AK3 is detected in all stages of parasite development in the fly (in the Accepted: June 29, 2015 midgut and in the salivary glands) as well as in bloodstream cells, but with predominance at Published: July 28, 2015 insect stages. -

Characterization of Nudix Hydrolases: a Utilitarian
CHARACTERIZATION OF NUDIX HYDROLASES: A UTILITARIAN SUPERFAMILY OF ENZYMES By Andres Hernandez de la Peña A dissertation submitted to the Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland August, 2015 Abstract The present work details the structural and enzymatic characterization of Nudix hydrolases from three different organisms – Bdellovibrio bacteriovorus, Mycobacterium tuberculosis, and Tetrahymena thermophila. Each of these Nudix enzymes presents unique questions about their physiological function within their organism which are answered with a combination of structural biology, genetic manipulation, enzyme kinetics, and a wide range of protein assays. We demonstrate that RenU, from M. tuberculosis, is part of Redox Homeostasis Control System (RHOCS), which senses and regulates NADH concentrations. This control systems involves two other proteins, the serine/threonine protein kinase G (pknG) and the L13 ribosomal subunits, without which the bacterium fails to evade lysosomal delivery and falls prey to the oxidative arsenal of the macrophage host. Bd-NDPSase, a Nudix enzyme encoded by the B. bacteriovorus gene BD3179, localizes to the periplasmic space of the bacterium and hydrolyses at least four nucleoside diphosphate sugars in vitro. Through atomic-resolution models from X-ray diffraction, we identified a motif that differentiates this hydrolase from the similar, but more substrate specific, ADP- ribose hydrolase from E. coli. Lastly, we show that Nud1p from T. thermophila is a member of the Ezl1p complex, the histone methyltransferase Polycomb Group homologue of this protozoan. With the use of in vitro enzymatic assays we show, in addition, that Nu1dp hydrolyses CoA preferentially over acetyl-CoA and other nucleoside derivatives. -

Experimental Validation of the Regulated Expression of Large Numbers of Non-Coding Rnas from the Mouse Genome
Downloaded from genome.cshlp.org on September 30, 2021 - Published by Cold Spring Harbor Laboratory Press Article Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome Timothy Ravasi,1,4,5 Harukazu Suzuki,2,4 Ken C. Pang,1,3,4 Shintaro Katayama,2,4 Masaaki Furuno,2,4,6 Rie Okunishi,2 Shiro Fukuda,2 Kelin Ru,1 Martin C. Frith,1,2 M. Milena Gongora,1 Sean M. Grimmond,1 David A. Hume,1 Yoshihide Hayashizaki,2 and John S. Mattick1,7 1ARC Special Research Centre for Functional and Applied Genomics, Institute for Molecular Bioscience, University of Queensland, Brisbane QLD 4072, Australia; 2Laboratory for Genome Exploration Research Group, RIKEN Genomic Science Center, RIKEN Yokohama Institute, Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan; 3T Cell Laboratory, Ludwig Institute for Cancer Research, Austin & Repatriation Medical Centre, Heidelberg VIC 3084, Australia Recent large-scale analyses of mainly full-length cDNA libraries generated from a variety of mouse tissues indicated that almost half of all representative cloned sequences did not contain an apparent protein-coding sequence, and were putatively derived from non-protein-coding RNA (ncRNA) genes. However, many of these clones were singletons and the majority were unspliced, raising the possibility that they may be derived from genomic DNA or unprocessed pre-mRNA contamination during library construction, or alternatively represent nonspecific “transcriptional noise.” Here we show, using reverse transcriptase-dependent PCR, microarray, and Northern blot analyses, that many of these clones were derived from genuine transcripts of unknown function whose expression appears to be regulated. -

RNF2 Antibody Order 021-34695924 [email protected] Support 400-6123-828 50Ul [email protected] 100 Ul √ √ Web
TD7403 RNF2 Antibody Order 021-34695924 [email protected] Support 400-6123-828 50ul [email protected] 100 uL √ √ Web www.ab-mart.com.cn Description: E3 ubiquitin-protein ligase that mediates monoubiquitination of 'Lys-119' of histone H2A (H2AK119Ub), thereby playing a central role in histone code and gene regulation. H2AK119Ub gives a specific tag for epigenetic transcriptional repression and participates in X chromosome inactivation of female mammals. May be involved in the initiation of both imprinted and random X inactivation (By similarity). Essential component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones, rendering chromatin heritably changed in its expressibility. E3 ubiquitin-protein ligase activity is enhanced by BMI1/PCGF4. Acts as the main E3 ubiquitin ligase on histone H2A of the PRC1 complex, while RING1 may rather act as a modulator of RNF2/RING2 activity (Probable). Association with the chromosomal DNA is cell-cycle dependent. In resting B- and T-lymphocytes, interaction with AURKB leads to block its activity, thereby maintaining transcription in resting lymphocytes (By similarity). Uniprot:Q99496 Alternative Names: BAP 1; BAP1; DING; DinG protein; E3 ubiquitin protein ligase RING 2; E3 ubiquitin protein ligase RING2; E3 ubiquitin-protein ligase RING2; HIP2 interacting protein 3; HIP2- interacting protein 3; HIPI 3; HIPI3; Huntingtin interacting protein 2 interacting protein 3; Huntingtin-interacting protein 2-interacting protein 3; OTTHUMP00000060668; Polycomb M33 interacting protein Ring 1B; Polycomb M33 interacting protein Ring1B; Protein DinG; RING 1B; RING 2; RING finger protein 1B; RING finger protein 2; RING finger protein BAP 1; RING finger protein BAP-1; RING finger protein BAP1; RING1b; RING2_HUMAN; RNF 2; Rnf2; Specificity: RNF2 Antibody detects endogenous levels of total RNF2. -
Product Data Sheet
For research purposes only, not for human use Product Data Sheet RNF2 siRNA (Human) Catalog # Source Reactivity Applications CRH4092 Synthetic H RNAi Description siRNA to inhibit RNF2 expression using RNA interference Specificity RNF2 siRNA (Human) is a target-specific 19-23 nt siRNA oligo duplexes designed to knock down gene expression. Form Lyophilized powder Gene Symbol RNF2 Alternative Names BAP1; DING; HIPI3; RING1B; E3 ubiquitin-protein ligase RING2; Huntingtin-interacting protein 2-interacting protein 3; HIP2-interacting protein 3; Protein DinG; RING finger protein 1B; RING1b; RING finger protein 2; RING finger protein BAP-1 Entrez Gene 6045 (Human) SwissProt Q99496 (Human) Purity > 97% Quality Control Oligonucleotide synthesis is monitored base by base through trityl analysis to ensure appropriate coupling efficiency. The oligo is subsequently purified by affinity-solid phase extraction. The annealed RNA duplex is further analyzed by mass spectrometry to verify the exact composition of the duplex. Each lot is compared to the previous lot by mass spectrometry to ensure maximum lot-to-lot consistency. Components We offers pre-designed sets of 3 different target-specific siRNA oligo duplexes of human RNF2 gene. Each vial contains 5 nmol of lyophilized siRNA. The duplexes can be transfected individually or pooled together to achieve knockdown of the target gene, which is most commonly assessed by qPCR or western blot. Our siRNA oligos Application key: E- ELISA, WB- Western blot, IH- Immunohistochemistry, IF- Immunofluorescence, FC- -

'Next- Generation' Sequencing Data Analysis
Novel Algorithm Development for ‘Next- Generation’ Sequencing Data Analysis Agne Antanaviciute Submitted in accordance with the requirements for the degree of Doctor of Philosophy University of Leeds School of Medicine Leeds Institute of Biomedical and Clinical Sciences 12/2017 ii The candidate confirms that the work submitted is her own, except where work which has formed part of jointly-authored publications has been included. The contribution of the candidate and the other authors to this work has been explicitly given within the thesis where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement ©2017 The University of Leeds and Agne Antanaviciute The right of Agne Antanaviciute to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. Acknowledgements I would like to thank all the people who have contributed to this work. First and foremost, my supervisors Dr Ian Carr, Professor David Bonthron and Dr Christopher Watson, who have provided guidance, support and motivation. I could not have asked for a better supervisory team. I would also like to thank my collaborators Dr Belinda Baquero and Professor Adrian Whitehouse for opening new, interesting research avenues. A special thanks to Dr Belinda Baquero for all the hard wet lab work without which at least half of this thesis would not exist. Thanks to everyone at the NGS Facility – Carolina Lascelles, Catherine Daley, Sally Harrison, Ummey Hany and Laura Crinnion – for the generation of NGS data used in this work and creating a supportive and stimulating work environment.