In Silico Prioritization of Transporter-Drug Relationships From
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Supplemental Figure 1. Vimentin
Double mutant specific genes Transcript gene_assignment Gene Symbol RefSeq FDR Fold- FDR Fold- FDR Fold- ID (single vs. Change (double Change (double Change wt) (single vs. wt) (double vs. single) (double vs. wt) vs. wt) vs. single) 10485013 BC085239 // 1110051M20Rik // RIKEN cDNA 1110051M20 gene // 2 E1 // 228356 /// NM 1110051M20Ri BC085239 0.164013 -1.38517 0.0345128 -2.24228 0.154535 -1.61877 k 10358717 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 /// BC 1700025G04Rik NM_197990 0.142593 -1.37878 0.0212926 -3.13385 0.093068 -2.27291 10358713 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 1700025G04Rik NM_197990 0.0655213 -1.71563 0.0222468 -2.32498 0.166843 -1.35517 10481312 NM_027283 // 1700026L06Rik // RIKEN cDNA 1700026L06 gene // 2 A3 // 69987 /// EN 1700026L06Rik NM_027283 0.0503754 -1.46385 0.0140999 -2.19537 0.0825609 -1.49972 10351465 BC150846 // 1700084C01Rik // RIKEN cDNA 1700084C01 gene // 1 H3 // 78465 /// NM_ 1700084C01Rik BC150846 0.107391 -1.5916 0.0385418 -2.05801 0.295457 -1.29305 10569654 AK007416 // 1810010D01Rik // RIKEN cDNA 1810010D01 gene // 7 F5 // 381935 /// XR 1810010D01Rik AK007416 0.145576 1.69432 0.0476957 2.51662 0.288571 1.48533 10508883 NM_001083916 // 1810019J16Rik // RIKEN cDNA 1810019J16 gene // 4 D2.3 // 69073 / 1810019J16Rik NM_001083916 0.0533206 1.57139 0.0145433 2.56417 0.0836674 1.63179 10585282 ENSMUST00000050829 // 2010007H06Rik // RIKEN cDNA 2010007H06 gene // --- // 6984 2010007H06Rik ENSMUST00000050829 0.129914 -1.71998 0.0434862 -2.51672 -

Phosphate Transporters of the SLC20 and SLC34 Families
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2013 Phosphate transporters of the SLC20 and SLC34 families Forster, Ian C ; Hernando, Nati ; Biber, Jürg ; Murer, Heini Abstract: Transport of inorganic phosphate (Pi) across the plasma membrane is essential for normal cellular function. Members of two families of SLC proteins (SLC20 and SLC34) act as Na(+)-dependent, secondary-active cotransporters to transport Pi across cell membranes. The SLC34 proteins are ex- pressed in specific organs important for Pi homeostasis: NaPi-IIa (SLC34A1) and NaPi-IIc (SLC34A3) fulfill essential roles in Pi reabsorption in the kidney proximal tubule and NaPi-IIb (SLC34A2) medi- ates Pi absorption in the gut. The SLC20 proteins, PiT-1 (SLC20A1), PiT-2 (SLC20A2) are expressed ubiquitously in all tissues and although generally considered as ”housekeeping” transport proteins, the discovery of tissue-specific activity, regulatory pathways and gene-related pathophysiologies, is redefining their importance. This review summarizes our current knowledge of SLC20 and SLC34 proteins in terms of their basic molecular characteristics, physiological roles, known pathophysiology and pharmacology. DOI: https://doi.org/10.1016/j.mam.2012.07.007 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-79439 Journal Article Accepted Version Originally published at: Forster, Ian C; Hernando, Nati; Biber, Jürg; Murer, Heini (2013). Phosphate transporters of the SLC20 and SLC34 families. Molecular Aspects of Medicine, 34(2-3):386-395. DOI: https://doi.org/10.1016/j.mam.2012.07.007 1 Mini-reviews Series-Molecular Aspects of Medicine (JMAM) Phosphate transporters of the SLC20 and SLC34 families Ian C. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

CD157 and CD200 at the Crossroads of Endothelial Remodeling and Immune Regulation
Editorial Commentary Page 1 of 7 CD157 and CD200 at the crossroads of endothelial remodeling and immune regulation Masuko Katoh1, Masaru Katoh2 1M & M PrecMed, Tokyo, Japan; 2Department of Omics Network, National Cancer Center, Tokyo, Japan Correspondence to: Masaru Katoh. Department of Omics Network, National Cancer Center, 5-1-1 Tsukiji, Chuo-ward, Tokyo 104-0045, Japan. Email: [email protected]. Comment on: Wakabayashi T, Naito H, Suehiro JI, et al. CD157 Marks Tissue-Resident Endothelial Stem Cells with Homeostatic and Regenerative Properties. Cell Stem Cell 2018;22:384-97.e6. Received: 11 March 2019; Accepted: 08 April 2019; Published: 19 April 2019. doi: 10.21037/sci.2019.04.01 View this article at: http://dx.doi.org/10.21037/sci.2019.04.01 The endothelial cells that coat the inner wall of blood cells formed more CD31 (PECAM1)-positive colonies than vessels are essential for the maintenance of the vascular CD200 single-positive or CD157/CD200 double-negative network, metabolic homeostasis and stem cell populations liver endothelial cells in vitro. The expression levels of Atf3, in tissue or tumor microenvironments (1-3). Angiogenesis Fosl2, Myc and Sox7 were significantly upregulated in the is defined as neovascular formation through the sprouting CD157/CD200 double-positive cells compared with the and proliferation of endothelial cells from preexisting CD200 single-positive or CD157/CD200 double-negative blood vessels. VEGF (VEGFA) and FGFs that transduce cells; however, the functions of these transcription factors signals through VEGFR2 and FGFRs, respectively, are in CD157/CD200 double-positive endothelial cells remain representative proangiogenic factors (4,5). -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Identification of a Novel Nucleobase-Ascorbate Transporter
bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Identification of a novel Nucleobase-Ascorbate 2 Transporter family member in fish and amphibians 3 Diogo Oliveira*,1, André M. Machado*,1, Tiago Cardoso1, Mónica Lopes-Marques1, L. Filipe 4 C. Castro1,2● and Raquel Ruivo1● 5 1CIIMAR – Interdisciplinary Centre of Marine and Environmental Research, U. Porto – University 6 of Porto, Porto, Portugal 7 2Department of Biology, Faculty of Sciences, U. Porto - University of Porto, Portugal 8 *Equal contribution 9 ●Corresponding authors at: CIIMAR, Terminal de Cruzeiros do Porto de Leixões, Av. General 10 Norton de Matos s/n, 4450-208 Matosinhos, Portugal. Tel.: +351 223 401 831 11 12 Running title: Novel uric acid transporter in fish and amphibians 13 14 15 bioRxiv preprint doi: https://doi.org/10.1101/287870; this version posted December 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 16 Abstract:Nucleobase-Ascorbate Transporter (NAT) family includes ascorbic acid, nucleobases 17 and uric acid transporters: with a broad evolutionary distribution. In vertebrates, four members 18 have been previously recognized, the ascorbate transporters Slc23a1 and Slc3a2, the nucleobase 19 transporter Slc23a4 and an orphan transporter Slc23a3. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Mouse Slc22a2 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Slc22a2 Knockout Project (CRISPR/Cas9) Objective: To create a Slc22a2 knockout Mouse model (C57BL/6N) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Slc22a2 gene (NCBI Reference Sequence: NM_013667 ; Ensembl: ENSMUSG00000040966 ) is located on Mouse chromosome 17. 11 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 11 (Transcript: ENSMUST00000046959). Exon 2 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a knockout allele are viable and fertile and display no obvious phenotypic abnormalities. No significant defects in the renal secretion of a model organic cation are observed. Exon 2 starts from about 25.02% of the coding region. Exon 2 covers 6.27% of the coding region. The size of effective KO region: ~104 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 11 Legends Exon of mouse Slc22a2 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. Tandem repeats are found in the dot plot matrix. The gRNA site is selected outside of these tandem repeats. -

Differential Expression of Hydroxyurea Transporters in Normal and Polycythemia Vera Hematopoietic Stem and Progenitor Cell Subpopulations
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2021 Differential expression of hydroxyurea transporters in normal and polycythemia vera hematopoietic stem and progenitor cell subpopulations Tan, Ge ; Meier-Abt, Fabienne Abstract: Polycythemia vera (PV) is a myeloproliferative neoplasm marked by hyperproliferation of the myeloid lineages and the presence of an activating JAK2 mutation. Hydroxyurea (HU) is a standard treat- ment for high-risk patients with PV. Because disease-driving mechanisms are thought to arise in PV stem cells, effective treatments should target primarily the stem cell compartment. We tested for theantipro- liferative effect of patient treatment with HU in fluorescence-activated cell sorting-isolated hematopoietic stem/multipotent progenitor cells (HSC/MPPs) and more committed erythroid progenitors (common myeloid/megakaryocyte-erythrocyte progenitors [CMP/MEPs]) in PV using RNA-sequencing and gene set enrichment analysis. HU treatment led to significant downregulation of gene sets associated with cell proliferation in PV HSCs/MPPs, but not in PV CMP/MEPs. To explore the mechanism underlying this finding, we assessed for expression of solute carrier membrane transporters, which mediate trans- membrane movement of drugs such as HU into target cells. The active HU uptake transporter OCTN1 was upregulated in HSC/MPPs compared with CMP/MEPs of untreated patients with PV, and the HU diffusion facilitator urea transporter B (UTB) was downregulated in HSC/MPPs compared withCM- P/MEPs in all patient and control groups tested. These findings indicate a higher accumulation ofHU within PV HSC/MPPs compared with PV CMP/MEPs and provide an explanation for the differential effects of HU in HSC/MPPs and CMP/MEPs of patients with PV. -
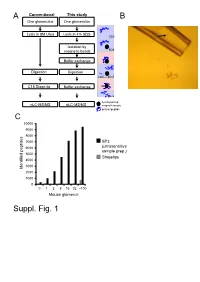
Suppl. Fig. 1 A
A Conventional This study B One glomerulus One glomerulus Lysis in 8M Urea Lysis in 4% SDS SDS Isolation by magnetic beads SDS Buffer exchange Digestion Digestion Try LysC pH>7 C18 Stage tip Buffer exchange pH<4 functionalized nLC-MS/MS nLC-MS/MS magnetic beads protein/peptide C 10000 9000 8000 7000 SP3 6000 (ultrasensitive sample prep.) 5000 Stagetips 4000 3000 Identified peptides 2000 1000 0 0 1 2 8 16 32 ~100 Mouse glomeruli Suppl. Fig. 1 S1. Sample preparation methods. A. Scheme of conventional (C18 Stage Tip) and ultrasensitive proteomic sample preparation methods. B. Manual microdissection and isolation of single glomeruli. The arrow indicates a single isolated glomerulus subjected to proteome analysis. The tip capacity is 10µl. C. Stagetips, a conventional proteomic sample preparation method, is compared with ultrasensitive (SP3) proteome analysis. The indicated amounts of mouse glomeruli were prepared by the respective sample preparation protocols, and peptide numbers after nLC-MS/MS are plotted (all FDR<0.01). A 100 1 Tubule B 80 60 40 Intensity CCD 20 40 0 100 20 S1 TAL 80 0 Tubule proximal tubule Loading... 60 0 Intensity 40 S2 20 -20 control Component 2 (22.5%) S3 0 0 10 20 30 40 50 60 -40 -20 0 20 40 Time (min) Component 2 (47.1%) C D 6000 Mouse proximal tubule 5000 8 4000 3000 7 (iBAQ) Loading... 10 2000 log 6 Number of peptides 1000 0 5 Control mouse mouse mouse human proximal mTAL CCD proximal 0 1500 tubule tubule Rank protein (S1) (S1) R=0.7 E F 8 ACTB Human proximal tubule 8 7 SLC9A3R1 ATP1A1 SLC3A2 SLC25A5 ATP1B1 7 SLC5A2 SLC25A3 CUBN CLTC SLC27A2 SLC5A12 LRP2 6 SLC25A13 SLC25A10 SLC5A1 SLC22A6 6 SLC25A4 Loading.. -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n