STATS 331 Introduction to Bayesian Statistics Brendon J. Brewer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -
3.3 Bayes' Formula
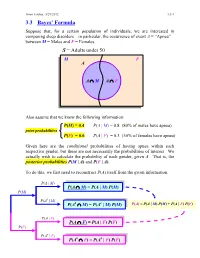
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 . -

The Bayesian Approach to Statistics
THE BAYESIAN APPROACH TO STATISTICS ANTHONY O’HAGAN INTRODUCTION the true nature of scientific reasoning. The fi- nal section addresses various features of modern By far the most widely taught and used statisti- Bayesian methods that provide some explanation for the rapid increase in their adoption since the cal methods in practice are those of the frequen- 1980s. tist school. The ideas of frequentist inference, as set out in Chapter 5 of this book, rest on the frequency definition of probability (Chapter 2), BAYESIAN INFERENCE and were developed in the first half of the 20th century. This chapter concerns a radically differ- We first present the basic procedures of Bayesian ent approach to statistics, the Bayesian approach, inference. which depends instead on the subjective defini- tion of probability (Chapter 3). In some respects, Bayesian methods are older than frequentist ones, Bayes’s Theorem and the Nature of Learning having been the basis of very early statistical rea- Bayesian inference is a process of learning soning as far back as the 18th century. Bayesian from data. To give substance to this statement, statistics as it is now understood, however, dates we need to identify who is doing the learning and back to the 1950s, with subsequent development what they are learning about. in the second half of the 20th century. Over that time, the Bayesian approach has steadily gained Terms and Notation ground, and is now recognized as a legitimate al- ternative to the frequentist approach. The person doing the learning is an individual This chapter is organized into three sections. -

Paradoxes and Priors in Bayesian Regression
Paradoxes and Priors in Bayesian Regression Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Agniva Som, B. Stat., M. Stat. Graduate Program in Statistics The Ohio State University 2014 Dissertation Committee: Dr. Christopher M. Hans, Advisor Dr. Steven N. MacEachern, Co-advisor Dr. Mario Peruggia c Copyright by Agniva Som 2014 Abstract The linear model has been by far the most popular and most attractive choice of a statistical model over the past century, ubiquitous in both frequentist and Bayesian literature. The basic model has been gradually improved over the years to deal with stronger features in the data like multicollinearity, non-linear or functional data pat- terns, violation of underlying model assumptions etc. One valuable direction pursued in the enrichment of the linear model is the use of Bayesian methods, which blend information from the data likelihood and suitable prior distributions placed on the unknown model parameters to carry out inference. This dissertation studies the modeling implications of many common prior distri- butions in linear regression, including the popular g prior and its recent ameliorations. Formalization of desirable characteristics for model comparison and parameter esti- mation has led to the growth of appropriate mixtures of g priors that conform to the seven standard model selection criteria laid out by Bayarri et al. (2012). The existence of some of these properties (or lack thereof) is demonstrated by examining the behavior of the prior under suitable limits on the likelihood or on the prior itself. -

A Widely Applicable Bayesian Information Criterion
JournalofMachineLearningResearch14(2013)867-897 Submitted 8/12; Revised 2/13; Published 3/13 A Widely Applicable Bayesian Information Criterion Sumio Watanabe [email protected] Department of Computational Intelligence and Systems Science Tokyo Institute of Technology Mailbox G5-19, 4259 Nagatsuta, Midori-ku Yokohama, Japan 226-8502 Editor: Manfred Opper Abstract A statistical model or a learning machine is called regular if the map taking a parameter to a prob- ability distribution is one-to-one and if its Fisher information matrix is always positive definite. If otherwise, it is called singular. In regular statistical models, the Bayes free energy, which is defined by the minus logarithm of Bayes marginal likelihood, can be asymptotically approximated by the Schwarz Bayes information criterion (BIC), whereas in singular models such approximation does not hold. Recently, it was proved that the Bayes free energy of a singular model is asymptotically given by a generalized formula using a birational invariant, the real log canonical threshold (RLCT), instead of half the number of parameters in BIC. Theoretical values of RLCTs in several statistical models are now being discovered based on algebraic geometrical methodology. However, it has been difficult to estimate the Bayes free energy using only training samples, because an RLCT depends on an unknown true distribution. In the present paper, we define a widely applicable Bayesian information criterion (WBIC) by the average log likelihood function over the posterior distribution with the inverse temperature 1/logn, where n is the number of training samples. We mathematically prove that WBIC has the same asymptotic expansion as the Bayes free energy, even if a statistical model is singular for or unrealizable by a statistical model. -

Part IV: Monte Carlo and Nonparametric Bayes Outline
Part IV: Monte Carlo and nonparametric Bayes Outline Monte Carlo methods Nonparametric Bayesian models Outline Monte Carlo methods Nonparametric Bayesian models The Monte Carlo principle • The expectation of f with respect to P can be approximated by 1 n E P(x)[ f (x)] " # f (xi ) n i=1 where the xi are sampled from P(x) • Example: the average # of spots on a die roll ! The Monte Carlo principle The law of large numbers n E P(x)[ f (x)] " # f (xi ) i=1 Average number of spots ! Number of rolls Two uses of Monte Carlo methods 1. For solving problems of probabilistic inference involved in developing computational models 2. As a source of hypotheses about how the mind might solve problems of probabilistic inference Making Bayesian inference easier P(d | h)P(h) P(h | d) = $P(d | h ") P(h ") h " # H Evaluating the posterior probability of a hypothesis requires considering all hypotheses ! Modern Monte Carlo methods let us avoid this Modern Monte Carlo methods • Sampling schemes for distributions with large state spaces known up to a multiplicative constant • Two approaches: – importance sampling (and particle filters) – Markov chain Monte Carlo Importance sampling Basic idea: generate from the wrong distribution, assign weights to samples to correct for this E p(x)[ f (x)] = " f (x)p(x)dx p(x) = f (x) q(x)dx " q(x) n ! 1 p(xi ) " # f (xi ) for xi ~ q(x) n i=1 q(xi ) ! ! Importance sampling works when sampling from proposal is easy, target is hard An alternative scheme… n 1 p(xi ) E p(x)[ f (x)] " # f (xi ) for xi ~ q(x) n i=1 q(xi ) n p(xi -

Marginal Likelihood
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! [email protected]! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 2 Last Class •" In our last class, we looked at: -" Statistical Decision Theory -" Linear Regression Models -" Linear Basis Function Models -" Regularized Linear Regression Models -" Bias-Variance Decomposition •" We will now look at the Bayesian framework and Bayesian Linear Regression Models. Bayesian Approach •" We formulate our knowledge about the world probabilistically: -" We define the model that expresses our knowledge qualitatively (e.g. independence assumptions, forms of distributions). -" Our model will have some unknown parameters. -" We capture our assumptions, or prior beliefs, about unknown parameters (e.g. range of plausible values) by specifying the prior distribution over those parameters before seeing the data. •" We observe the data. •" We compute the posterior probability distribution for the parameters, given observed data. •" We use this posterior distribution to: -" Make predictions by averaging over the posterior distribution -" Examine/Account for uncertainly in the parameter values. -" Make decisions by minimizing expected posterior loss. (See Radford Neal’s NIPS tutorial on ``Bayesian Methods for Machine Learning'’) Posterior Distribution •" The posterior distribution for the model parameters can be found by combining the prior with the likelihood for the parameters given the data. •" This is accomplished using Bayes’ -

Naïve Bayes Classifier
NAÏVE BAYES CLASSIFIER Professor Tom Fomby Department of Economics Southern Methodist University Dallas, Texas 75275 April 2008 The Naïve Bayes classifier is a classification method based on Bayes Theorem. Let C j denote that an output belongs to the j-th class, j 1,2, J out of J possible classes. Let P(C j | X1, X 2 ,, X p ) denote the (posterior) probability of belonging in the j-th class given the individual characteristics X1, X 2 ,, X p . Furthermore, let P(X1, X 2 ,, X p | C j )denote the probability of a case with individual characteristics belonging to the j-th class and P(C j ) denote the unconditional (i.e. without regard to individual characteristics) prior probability of belonging to the j-th class. For a total of J classes, Bayes theorem gives us the following probability rule for calculating the case-specific probability of falling into the j-th class: P(X , X ,, X | C ) P(C ) P(C | X , X ,, X ) 1 2 p j j (1) j 1 2 p Denom where Denom P(X1, X 2 ,, X p | C1 )P(C1 ) P(X1, X 2 ,, X p | CJ )P(CJ ) . Of course the conditional class probabilities of (1) are exhaustive in that a case J (X1, X 2 ,, X p ) has to fall in one of the J cases. That is, P(C j | X1 , X 2 ,, X p ) 1. j1 The difficulty with using (1) is that in situations where the number of cases (X1, X 2 ,, X p ) is few and distinct and the number of classes J is large, there may be many instances where the probabilities of cases falling in specific classes, , are frequently equal to zero for the majority of classes. -

Constraints Versus Priors † Philip B
SIAM/ASA J. UNCERTAINTY QUANTIFICATION c 2015 Society for Industrial and Applied Mathematics Vol. 3, pp. 586–598 and American Statistical Association ∗ Constraints versus Priors † Philip B. Stark Abstract. There are deep and important philosophical differences between Bayesian and frequentist approaches to quantifying uncertainty. However, some practitioners choose between these approaches primar- ily on the basis of convenience. For instance, the ability to incorporate parameter constraints is sometimes cited as a reason to use Bayesian methods. This reflects two misunderstandings: First, frequentist methods can indeed incorporate constraints on parameter values. Second, it ignores the crucial question of what the result of the analysis will mean. Bayesian and frequentist measures of uncertainty have similar sounding names but quite different meanings. For instance, Bayesian uncertainties typically involve expectations with respect to the posterior distribution of the param- eter, holding the data fixed; frequentist uncertainties typically involve expectations with respect to the distribution of the data, holding the parameter fixed. Bayesian methods, including methods incorporating parameter constraints, require supplementing the constraints with a prior probability distribution for parameter values. This can cause frequentist and Bayesian estimates and their nom- inal uncertainties to differ substantially, even when the prior is “uninformative.” This paper gives simple examples where “uninformative” priors are, in fact, extremely informative, and sketches how to measure how much information the prior adds to the constraint. Bayesian methods can have good frequentist behavior, and a frequentist can use Bayesian methods and quantify the uncertainty by frequentist means—but absent a meaningful prior, Bayesian uncertainty measures lack meaning. The paper ends with brief reflections on practice. -

9 Introduction to Hierarchical Models
9 Introduction to Hierarchical Models One of the important features of a Bayesian approach is the relative ease with which hierarchical models can be constructed and estimated using Gibbs sampling. In fact, one of the key reasons for the recent growth in the use of Bayesian methods in the social sciences is that the use of hierarchical models has also increased dramatically in the last two decades. Hierarchical models serve two purposes. One purpose is methodological; the other is substantive. Methodologically, when units of analysis are drawn from clusters within a population (communities, neighborhoods, city blocks, etc.), they can no longer be considered independent. Individuals who come from the same cluster will be more similar to each other than they will be to individuals from other clusters. Therefore, unobserved variables may in- duce statistical dependence between observations within clusters that may be uncaptured by covariates within the model, violating a key assumption of maximum likelihood estimation as it is typically conducted when indepen- dence of errors is assumed. Recall that a likelihood function, when observations are independent, is simply the product of the density functions for each ob- servation taken over all the observations. However, when independence does not hold, we cannot construct the likelihood as simply. Thus, one reason for constructing hierarchical models is to compensate for the biases—largely in the standard errors—that are introduced when the independence assumption is violated. See Ezell, Land, and Cohen (2003) for a thorough review of the approaches that have been used to correct standard errors in hazard model- ing applications with repeated events, one class of models in which repeated measurement yields hierarchical clustering. -

Bayesian Model Selection
Bayesian model selection Consider the regression problem, where we want to predict the values of an unknown function d N y : R ! R given examples D = (xi; yi) i=1 to serve as training data. In Bayesian linear regression, we made the following assumption about y(x): y(x) = φ(x)>w + "(x); (1) where φ(x) is a now explicitly-written feature expansion of x. We proceed in the normal Bayesian way: we place Gaussian priors on our unknowns, the parameters w and the residuals ", then derive the posterior distribution over w given D, which we use to make predictions. One question left unanswered is how to choose a good feature expansion function φ(x). For example, a purely linear model could use φ(x) = [1; x]>, whereas a quadratic model could use φ(x) = [1; x; x2]>, etc. In general, arbitrary feature expansions φ are allowed. How can I select between them? Even more generally, how do I select whether I should use linear regression or a completely dierent probabilistic model to explain my data? These are questions of model selection, and naturally there is a Bayesian approach to it. Before we continue our discussion of model selection, we will rst dene the word model, which is often used loosely without explicit denition. A model is a parametric family of probability distributions, each of which can explain the observed data. Another way to explain the concept of a model is that if we have chosen a likelihood p(D j θ) for our data, which depends on a parameter θ, then the model is the set of all likelihoods (each one of which is a distribution over D) for every possible value of the parameter θ.