Making Models with Bayes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Statistical Methods for Data Science, Lecture 5 Interval Estimates; Comparing Systems
Statistical methods for Data Science, Lecture 5 Interval estimates; comparing systems Richard Johansson November 18, 2018 statistical inference: overview I estimate the value of some parameter (last lecture): I what is the error rate of my drug test? I determine some interval that is very likely to contain the true value of the parameter (today): I interval estimate for the error rate I test some hypothesis about the parameter (today): I is the error rate significantly different from 0.03? I are users significantly more satisfied with web page A than with web page B? -20pt “recipes” I in this lecture, we’ll look at a few “recipes” that you’ll use in the assignment I interval estimate for a proportion (“heads probability”) I comparing a proportion to a specified value I comparing two proportions I additionally, we’ll see the standard method to compute an interval estimate for the mean of a normal I I will also post some pointers to additional tests I remember to check that the preconditions are satisfied: what kind of experiment? what assumptions about the data? -20pt overview interval estimates significance testing for the accuracy comparing two classifiers p-value fishing -20pt interval estimates I if we get some estimate by ML, can we say something about how reliable that estimate is? I informally, an interval estimate for the parameter p is an interval I = [plow ; phigh] so that the true value of the parameter is “likely” to be contained in I I for instance: with 95% probability, the error rate of the spam filter is in the interval [0:05; 0:08] -20pt frequentists and Bayesians again. -

DSA 5403 Bayesian Statistics: an Advancing Introduction 16 Units – Each Unit a Week’S Work
DSA 5403 Bayesian Statistics: An Advancing Introduction 16 units – each unit a week’s work. The following are the contents of the course divided into chapters of the book Doing Bayesian Data Analysis . by John Kruschke. The course is structured around the above book but will be embellished with more theoretical content as needed. The book can be obtained from the library : http://www.sciencedirect.com/science/book/9780124058880 Topics: This is divided into three parts From the book: “Part I The basics: models, probability, Bayes' rule and r: Introduction: credibility, models, and parameters; The R programming language; What is this stuff called probability?; Bayes' rule Part II All the fundamentals applied to inferring a binomial probability: Inferring a binomial probability via exact mathematical analysis; Markov chain Monte C arlo; J AG S ; Hierarchical models; Model comparison and hierarchical modeling; Null hypothesis significance testing; Bayesian approaches to testing a point ("Null") hypothesis; G oals, power, and sample size; S tan Part III The generalized linear model: Overview of the generalized linear model; Metric-predicted variable on one or two groups; Metric predicted variable with one metric predictor; Metric predicted variable with multiple metric predictors; Metric predicted variable with one nominal predictor; Metric predicted variable with multiple nominal predictors; Dichotomous predicted variable; Nominal predicted variable; Ordinal predicted variable; C ount predicted variable; Tools in the trunk” Part 1: The Basics: Models, Probability, Bayes’ Rule and R Unit 1 Chapter 1, 2 Credibility, Models, and Parameters, • Derivation of Bayes’ rule • Re-allocation of probability • The steps of Bayesian data analysis • Classical use of Bayes’ rule o Testing – false positives etc o Definitions of prior, likelihood, evidence and posterior Unit 2 Chapter 3 The R language • Get the software • Variables types • Loading data • Functions • Plots Unit 3 Chapter 4 Probability distributions. -

An Introduction to Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A
NESUG 2010 Statistics and Analysis An Animated Guide: An Introduction To Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A. ABSTRACT: This paper will be a brief introduction to Poisson regression (theory, steps to be followed, complications and interpretation) via a worked example. It is hoped that this will increase motivation towards learning this useful statistical technique. INTRODUCTION: Poisson regression is available in SAS through the GENMOD procedure (generalized modeling). It is appropriate when: 1) the process that generates the conditional Y distributions would, theoretically, be expected to be a Poisson random process and 2) when there is no evidence of overdispersion and 3) when the mean of the marginal distribution is less than ten (preferably less than five and ideally close to one). THE POISSON DISTRIBUTION: The Poison distribution is a discrete Percent of observations where the random variable X is expected distribution and is appropriate for to have the value x, given that the Poisson distribution has a mean modeling counts of observations. of λ= P(X=x, λ ) = (e - λ * λ X) / X! Counts are observed cases, like the 0.4 count of measles cases in cities. You λ can simply model counts if all data 0.35 were collected in the same measuring 0.3 unit (e.g. the same number of days or 0.3 0.5 0.8 same number of square feet). 0.25 λ 1 0.2 3 You can use the Poisson Distribution = 5 for modeling rates (rates are counts 0.15 20 per unit) if the units of collection were 8 different. -

Generalized Linear Models (Glms)
San Jos´eState University Math 261A: Regression Theory & Methods Generalized Linear Models (GLMs) Dr. Guangliang Chen This lecture is based on the following textbook sections: • Chapter 13: 13.1 – 13.3 Outline of this presentation: • What is a GLM? • Logistic regression • Poisson regression Generalized Linear Models (GLMs) What is a GLM? In ordinary linear regression, we assume that the response is a linear function of the regressors plus Gaussian noise: 0 2 y = β0 + β1x1 + ··· + βkxk + ∼ N(x β, σ ) | {z } |{z} linear form x0β N(0,σ2) noise The model can be reformulate in terms of • distribution of the response: y | x ∼ N(µ, σ2), and • dependence of the mean on the predictors: µ = E(y | x) = x0β Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University3/24 Generalized Linear Models (GLMs) beta=(1,2) 5 4 3 β0 + β1x b y 2 y 1 0 −1 0.0 0.2 0.4 0.6 0.8 1.0 x x Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University4/24 Generalized Linear Models (GLMs) Generalized linear models (GLM) extend linear regression by allowing the response variable to have • a general distribution (with mean µ = E(y | x)) and • a mean that depends on the predictors through a link function g: That is, g(µ) = β0x or equivalently, µ = g−1(β0x) Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University5/24 Generalized Linear Models (GLMs) In GLM, the response is typically assumed to have a distribution in the exponential family, which is a large class of probability distributions that have pdfs of the form f(x | θ) = a(x)b(θ) exp(c(θ) · T (x)), including • Normal - ordinary linear regression • Bernoulli - Logistic regression, modeling binary data • Binomial - Multinomial logistic regression, modeling general cate- gorical data • Poisson - Poisson regression, modeling count data • Exponential, Gamma - survival analysis Dr. -

Assessing Fairness with Unlabeled Data and Bayesian Inference
Can I Trust My Fairness Metric? Assessing Fairness with Unlabeled Data and Bayesian Inference Disi Ji1 Padhraic Smyth1 Mark Steyvers2 1Department of Computer Science 2Department of Cognitive Sciences University of California, Irvine [email protected] [email protected] [email protected] Abstract We investigate the problem of reliably assessing group fairness when labeled examples are few but unlabeled examples are plentiful. We propose a general Bayesian framework that can augment labeled data with unlabeled data to produce more accurate and lower-variance estimates compared to methods based on labeled data alone. Our approach estimates calibrated scores for unlabeled examples in each group using a hierarchical latent variable model conditioned on labeled examples. This in turn allows for inference of posterior distributions with associated notions of uncertainty for a variety of group fairness metrics. We demonstrate that our approach leads to significant and consistent reductions in estimation error across multiple well-known fairness datasets, sensitive attributes, and predictive models. The results show the benefits of using both unlabeled data and Bayesian inference in terms of assessing whether a prediction model is fair or not. 1 Introduction Machine learning models are increasingly used to make important decisions about individuals. At the same time it has become apparent that these models are susceptible to producing systematically biased decisions with respect to sensitive attributes such as gender, ethnicity, and age [Angwin et al., 2017, Berk et al., 2018, Corbett-Davies and Goel, 2018, Chen et al., 2019, Beutel et al., 2019]. This has led to a significant amount of recent work in machine learning addressing these issues, including research on both (i) definitions of fairness in a machine learning context (e.g., Dwork et al. -

Bayesian Inference Chapter 4: Regression and Hierarchical Models
Bayesian Inference Chapter 4: Regression and Hierarchical Models Conchi Aus´ınand Mike Wiper Department of Statistics Universidad Carlos III de Madrid Master in Business Administration and Quantitative Methods Master in Mathematical Engineering Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 1 / 35 Objective AFM Smith Dennis Lindley We analyze the Bayesian approach to fitting normal and generalized linear models and introduce the Bayesian hierarchical modeling approach. Also, we study the modeling and forecasting of time series. Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 2 / 35 Contents 1 Normal linear models 1.1. ANOVA model 1.2. Simple linear regression model 2 Generalized linear models 3 Hierarchical models 4 Dynamic models Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 3 / 35 Normal linear models A normal linear model is of the following form: y = Xθ + ; 0 where y = (y1;:::; yn) is the observed data, X is a known n × k matrix, called 0 the design matrix, θ = (θ1; : : : ; θk ) is the parameter set and follows a multivariate normal distribution. Usually, it is assumed that: 1 ∼ N 0 ; I : k φ k A simple example of normal linear model is the simple linear regression model T 1 1 ::: 1 where X = and θ = (α; β)T . x1 x2 ::: xn Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 4 / 35 Normal linear models Consider a normal linear model, y = Xθ + . A conjugate prior distribution is a normal-gamma distribution: -

Linear Mixed-Effects Modeling in SPSS: an Introduction to the MIXED Procedure
Technical report Linear Mixed-Effects Modeling in SPSS: An Introduction to the MIXED Procedure Table of contents Introduction. 1 Data preparation for MIXED . 1 Fitting fixed-effects models . 4 Fitting simple mixed-effects models . 7 Fitting mixed-effects models . 13 Multilevel analysis . 16 Custom hypothesis tests . 18 Covariance structure selection. 19 Random coefficient models . 20 Estimated marginal means. 25 References . 28 About SPSS Inc. 28 SPSS is a registered trademark and the other SPSS products named are trademarks of SPSS Inc. All other names are trademarks of their respective owners. © 2005 SPSS Inc. All rights reserved. LMEMWP-0305 Introduction The linear mixed-effects models (MIXED) procedure in SPSS enables you to fit linear mixed-effects models to data sampled from normal distributions. Recent texts, such as those by McCulloch and Searle (2000) and Verbeke and Molenberghs (2000), comprehensively review mixed-effects models. The MIXED procedure fits models more general than those of the general linear model (GLM) procedure and it encompasses all models in the variance components (VARCOMP) procedure. This report illustrates the types of models that MIXED handles. We begin with an explanation of simple models that can be fitted using GLM and VARCOMP, to show how they are translated into MIXED. We then proceed to fit models that are unique to MIXED. The major capabilities that differentiate MIXED from GLM are that MIXED handles correlated data and unequal variances. Correlated data are very common in such situations as repeated measurements of survey respondents or experimental subjects. MIXED extends repeated measures models in GLM to allow an unequal number of repetitions. -

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -

Multilevel and Mixed-Effects Modeling 391
386 Statistics with Stata , . d itl 770/ ""01' ncdctemp However the residuals pass tests for white noise 111uahtemp, compar e WI 1 /0 l' r : , , '12 19) (p = .65), and a plot of observed and predicted values shows a good visual fit (Figure . , Multilevel and Mixed-Effects predict uahhat2 solar, ENSO & C02" label variable uahhat2 "predicted from volcanoes, Modeling predict uahres2, resid label variable uahres2 "UAH residuals from ARMAX model" wntestq uahres2, lags(25) portmanteau test for white noise portmanteau (Q)statistic 21. 7197 Mixed-effects modeling is basically regression analysis allowing two kinds of effects: fixed =rob > chi2(25) 0.6519 effects, meaning intercepts and slopes meant to describe the population as a whole, just as in Figure 12.19 ordinary regression; and also random effects, meaning intercepts and slopes that can vary across subgroups of the sample. All of the regression-type methods shown so far in this book involve fixed effects only. Mixed-effects modeling opens a new range of possibilities for multilevel o models, growth curve analysis, and panel data or cross-sectional time series, "r~ 00 01 Albright and Marinova (2010) provide a practical comparison of mixed-modeling procedures uj"'! > found in Stata, SAS, SPSS and R with the hierarchical linear modeling (HLM) software 0>- m developed by Raudenbush and Bryck (2002; also Raudenbush et al. 2005). More detailed ~o c explanation of mixed modeling and its correspondences with HLM can be found in Rabe• ro (I! Hesketh and Skrondal (2012). Briefly, HLM approaches multilevel modeling in several steps, ::IN co I' specifying separate equations (for example) for levelland level 2 effects. -

Generalized Linear Models with Poisson Family: Applications in Ecology
UNIVERSITY OF ABOMEY- CALAVI *********** FACULTY OF AGRONOMIC SCIENCES *************** **************** Master Program in Statistics, Major Biostatistics 1st batch Generalized linear models with Poisson family: applications in ecology A thesis submitted to the Faculty of Agronomic Sciences in partial fulfillment of the requirements for the degree of the Master of Sciences in Biostatistics Presented by: LOKONON Enagnon Bruno Supervisor: Pr Romain L. GLELE KAKAÏ, Professor of Biostatistics and Forest estimation Academic year: 2014-2015 UNIVERSITE D’ABOMEY- CALAVI *********** FACULTE DES SCIENCES AGRONOMIQUES *************** ************** Programme de Master en Biostatistiques 1ère Promotion Modèles linéaires généralisés de la famille de Poisson : applications en écologie Mémoire soumis à la Faculté des Sciences Agronomiques pour obtenir le Diplôme de Master recherche en Biostatistiques Présenté par: LOKONON Enagnon Bruno Superviseur: Pr Romain L. GLELE KAKAÏ, Professeur titulaire de Biostatistiques et estimation forestière Année académique: 2014-2015 Certification I certify that this work has been achieved by LOKONON E. Bruno under my entire supervision at the University of Abomey-Calavi (Benin) in order to obtain his Master of Science degree in Biostatistics. Pr Romain L. GLELE KAKAÏ Professor of Biostatistics and Forest estimation i Acknowledgements This research was supported by WAAPP/PPAAO-BENIN (West African Agricultural Productivity Program/ Programme de Productivité Agricole en Afrique de l‟Ouest). This dissertation could only have been possible through the generous contributions of many people. First and foremost, I am grateful to my supervisor Pr Romain L. GLELE KAKAÏ, Professor of Biostatistics and Forest estimation who tirelessly played key role in orientation, scientific writing and mentoring during this research. In particular, I thank him for his prompt availability whenever needed. -
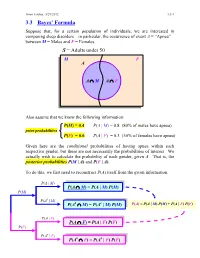
3.3 Bayes' Formula
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 .