Bayesian Model Selection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -
![Arxiv:1803.00360V2 [Stat.CO] 2 Mar 2018 Prone to Misinterpretation [2, 3]](https://docslib.b-cdn.net/cover/7623/arxiv-1803-00360v2-stat-co-2-mar-2018-prone-to-misinterpretation-2-3-297623.webp)
Arxiv:1803.00360V2 [Stat.CO] 2 Mar 2018 Prone to Misinterpretation [2, 3]
Computing Bayes factors to measure evidence from experiments: An extension of the BIC approximation Thomas J. Faulkenberry∗ Tarleton State University Bayesian inference affords scientists with powerful tools for testing hypotheses. One of these tools is the Bayes factor, which indexes the extent to which support for one hypothesis over another is updated after seeing the data. Part of the hesitance to adopt this approach may stem from an unfamiliarity with the computational tools necessary for computing Bayes factors. Previous work has shown that closed form approximations of Bayes factors are relatively easy to obtain for between-groups methods, such as an analysis of variance or t-test. In this paper, I extend this approximation to develop a formula for the Bayes factor that directly uses infor- mation that is typically reported for ANOVAs (e.g., the F ratio and degrees of freedom). After giving two examples of its use, I report the results of simulations which show that even with minimal input, this approximate Bayes factor produces similar results to existing software solutions. Note: to appear in Biometrical Letters. I. INTRODUCTION A. The Bayes factor Bayesian inference is a method of measurement that is Hypothesis testing is the primary tool for statistical based on the computation of P (H j D), which is called inference across much of the biological and behavioral the posterior probability of a hypothesis H, given data D. sciences. As such, most scientists are trained in classical Bayes' theorem casts this probability as null hypothesis significance testing (NHST). The scenario for testing a hypothesis is likely familiar to most readers of this journal. -
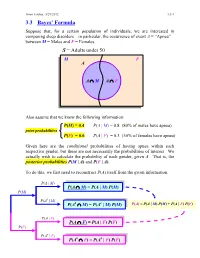
3.3 Bayes' Formula
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Naïve Bayes Classifier
CS 60050 Machine Learning Naïve Bayes Classifier Some slides taken from course materials of Tan, Steinbach, Kumar Bayes Classifier ● A probabilistic framework for solving classification problems ● Approach for modeling probabilistic relationships between the attribute set and the class variable – May not be possible to certainly predict class label of a test record even if it has identical attributes to some training records – Reason: noisy data or presence of certain factors that are not included in the analysis Probability Basics ● P(A = a, C = c): joint probability that random variables A and C will take values a and c respectively ● P(A = a | C = c): conditional probability that A will take the value a, given that C has taken value c P(A,C) P(C | A) = P(A) P(A,C) P(A | C) = P(C) Bayes Theorem ● Bayes theorem: P(A | C)P(C) P(C | A) = P(A) ● P(C) known as the prior probability ● P(C | A) known as the posterior probability Example of Bayes Theorem ● Given: – A doctor knows that meningitis causes stiff neck 50% of the time – Prior probability of any patient having meningitis is 1/50,000 – Prior probability of any patient having stiff neck is 1/20 ● If a patient has stiff neck, what’s the probability he/she has meningitis? P(S | M )P(M ) 0.5×1/50000 P(M | S) = = = 0.0002 P(S) 1/ 20 Bayesian Classifiers ● Consider each attribute and class label as random variables ● Given a record with attributes (A1, A2,…,An) – Goal is to predict class C – Specifically, we want to find the value of C that maximizes P(C| A1, A2,…,An ) ● Can we estimate -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 . -

The Bayesian Approach to Statistics
THE BAYESIAN APPROACH TO STATISTICS ANTHONY O’HAGAN INTRODUCTION the true nature of scientific reasoning. The fi- nal section addresses various features of modern By far the most widely taught and used statisti- Bayesian methods that provide some explanation for the rapid increase in their adoption since the cal methods in practice are those of the frequen- 1980s. tist school. The ideas of frequentist inference, as set out in Chapter 5 of this book, rest on the frequency definition of probability (Chapter 2), BAYESIAN INFERENCE and were developed in the first half of the 20th century. This chapter concerns a radically differ- We first present the basic procedures of Bayesian ent approach to statistics, the Bayesian approach, inference. which depends instead on the subjective defini- tion of probability (Chapter 3). In some respects, Bayesian methods are older than frequentist ones, Bayes’s Theorem and the Nature of Learning having been the basis of very early statistical rea- Bayesian inference is a process of learning soning as far back as the 18th century. Bayesian from data. To give substance to this statement, statistics as it is now understood, however, dates we need to identify who is doing the learning and back to the 1950s, with subsequent development what they are learning about. in the second half of the 20th century. Over that time, the Bayesian approach has steadily gained Terms and Notation ground, and is now recognized as a legitimate al- ternative to the frequentist approach. The person doing the learning is an individual This chapter is organized into three sections. -

LINEAR REGRESSION MODELS Likelihood and Reference Bayesian
{ LINEAR REGRESSION MODELS { Likeliho o d and Reference Bayesian Analysis Mike West May 3, 1999 1 Straight Line Regression Mo dels We b egin with the simple straight line regression mo del y = + x + i i i where the design p oints x are xed in advance, and the measurement/sampling i 2 errors are indep endent and normally distributed, N 0; for each i = i i 1;::: ;n: In this context, wehavelooked at general mo delling questions, data and the tting of least squares estimates of and : Now we turn to more formal likeliho o d and Bayesian inference. 1.1 Likeliho o d and MLEs The formal parametric inference problem is a multi-parameter problem: we 2 require inferences on the three parameters ; ; : The likeliho o d function has a simple enough form, as wenow show. Throughout, we do not indicate the design p oints in conditioning statements, though they are implicitly conditioned up on. Write Y = fy ;::: ;y g and X = fx ;::: ;x g: Given X and the mo del 1 n 1 n parameters, each y is the corresp onding zero-mean normal random quantity i plus the term + x ; so that y is normal with this term as its mean and i i i 2 variance : Also, since the are indep endent, so are the y : Thus i i 2 2 y j ; ; N + x ; i i with conditional density function 2 2 2 2 1=2 py j ; ; = expfy x =2 g=2 i i i for each i: Also, by indep endence, the joint density function is n Y 2 2 : py j ; ; = pY j ; ; i i=1 1 Given the observed resp onse values Y this provides the likeliho o d function for the three parameters: the joint density ab ove evaluated at the observed and 2 hence now xed values of Y; now viewed as a function of ; ; as they vary across p ossible parameter values. -

Paradoxes and Priors in Bayesian Regression
Paradoxes and Priors in Bayesian Regression Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Agniva Som, B. Stat., M. Stat. Graduate Program in Statistics The Ohio State University 2014 Dissertation Committee: Dr. Christopher M. Hans, Advisor Dr. Steven N. MacEachern, Co-advisor Dr. Mario Peruggia c Copyright by Agniva Som 2014 Abstract The linear model has been by far the most popular and most attractive choice of a statistical model over the past century, ubiquitous in both frequentist and Bayesian literature. The basic model has been gradually improved over the years to deal with stronger features in the data like multicollinearity, non-linear or functional data pat- terns, violation of underlying model assumptions etc. One valuable direction pursued in the enrichment of the linear model is the use of Bayesian methods, which blend information from the data likelihood and suitable prior distributions placed on the unknown model parameters to carry out inference. This dissertation studies the modeling implications of many common prior distri- butions in linear regression, including the popular g prior and its recent ameliorations. Formalization of desirable characteristics for model comparison and parameter esti- mation has led to the growth of appropriate mixtures of g priors that conform to the seven standard model selection criteria laid out by Bayarri et al. (2012). The existence of some of these properties (or lack thereof) is demonstrated by examining the behavior of the prior under suitable limits on the likelihood or on the prior itself. -

1 Estimation and Beyond in the Bayes Universe
ISyE8843A, Brani Vidakovic Handout 7 1 Estimation and Beyond in the Bayes Universe. 1.1 Estimation No Bayes estimate can be unbiased but Bayesians are not upset! No Bayes estimate with respect to the squared error loss can be unbiased, except in a trivial case when its Bayes’ risk is 0. Suppose that for a proper prior ¼ the Bayes estimator ±¼(X) is unbiased, Xjθ (8θ)E ±¼(X) = θ: This implies that the Bayes risk is 0. The Bayes risk of ±¼(X) can be calculated as repeated expectation in two ways, θ Xjθ 2 X θjX 2 r(¼; ±¼) = E E (θ ¡ ±¼(X)) = E E (θ ¡ ±¼(X)) : Thus, conveniently choosing either EθEXjθ or EX EθjX and using the properties of conditional expectation we have, θ Xjθ 2 θ Xjθ X θjX X θjX 2 r(¼; ±¼) = E E θ ¡ E E θ±¼(X) ¡ E E θ±¼(X) + E E ±¼(X) θ Xjθ 2 θ Xjθ X θjX X θjX 2 = E E θ ¡ E θ[E ±¼(X)] ¡ E ±¼(X)E θ + E E ±¼(X) θ Xjθ 2 θ X X θjX 2 = E E θ ¡ E θ ¢ θ ¡ E ±¼(X)±¼(X) + E E ±¼(X) = 0: Bayesians are not upset. To check for its unbiasedness, the Bayes estimator is averaged with respect to the model measure (Xjθ), and one of the Bayesian commandments is: Thou shall not average with respect to sample space, unless you have Bayesian design in mind. Even frequentist agree that insisting on unbiasedness can lead to bad estimators, and that in their quest to minimize the risk by trading off between variance and bias-squared a small dosage of bias can help. -

A Widely Applicable Bayesian Information Criterion
JournalofMachineLearningResearch14(2013)867-897 Submitted 8/12; Revised 2/13; Published 3/13 A Widely Applicable Bayesian Information Criterion Sumio Watanabe [email protected] Department of Computational Intelligence and Systems Science Tokyo Institute of Technology Mailbox G5-19, 4259 Nagatsuta, Midori-ku Yokohama, Japan 226-8502 Editor: Manfred Opper Abstract A statistical model or a learning machine is called regular if the map taking a parameter to a prob- ability distribution is one-to-one and if its Fisher information matrix is always positive definite. If otherwise, it is called singular. In regular statistical models, the Bayes free energy, which is defined by the minus logarithm of Bayes marginal likelihood, can be asymptotically approximated by the Schwarz Bayes information criterion (BIC), whereas in singular models such approximation does not hold. Recently, it was proved that the Bayes free energy of a singular model is asymptotically given by a generalized formula using a birational invariant, the real log canonical threshold (RLCT), instead of half the number of parameters in BIC. Theoretical values of RLCTs in several statistical models are now being discovered based on algebraic geometrical methodology. However, it has been difficult to estimate the Bayes free energy using only training samples, because an RLCT depends on an unknown true distribution. In the present paper, we define a widely applicable Bayesian information criterion (WBIC) by the average log likelihood function over the posterior distribution with the inverse temperature 1/logn, where n is the number of training samples. We mathematically prove that WBIC has the same asymptotic expansion as the Bayes free energy, even if a statistical model is singular for or unrealizable by a statistical model. -

Part IV: Monte Carlo and Nonparametric Bayes Outline
Part IV: Monte Carlo and nonparametric Bayes Outline Monte Carlo methods Nonparametric Bayesian models Outline Monte Carlo methods Nonparametric Bayesian models The Monte Carlo principle • The expectation of f with respect to P can be approximated by 1 n E P(x)[ f (x)] " # f (xi ) n i=1 where the xi are sampled from P(x) • Example: the average # of spots on a die roll ! The Monte Carlo principle The law of large numbers n E P(x)[ f (x)] " # f (xi ) i=1 Average number of spots ! Number of rolls Two uses of Monte Carlo methods 1. For solving problems of probabilistic inference involved in developing computational models 2. As a source of hypotheses about how the mind might solve problems of probabilistic inference Making Bayesian inference easier P(d | h)P(h) P(h | d) = $P(d | h ") P(h ") h " # H Evaluating the posterior probability of a hypothesis requires considering all hypotheses ! Modern Monte Carlo methods let us avoid this Modern Monte Carlo methods • Sampling schemes for distributions with large state spaces known up to a multiplicative constant • Two approaches: – importance sampling (and particle filters) – Markov chain Monte Carlo Importance sampling Basic idea: generate from the wrong distribution, assign weights to samples to correct for this E p(x)[ f (x)] = " f (x)p(x)dx p(x) = f (x) q(x)dx " q(x) n ! 1 p(xi ) " # f (xi ) for xi ~ q(x) n i=1 q(xi ) ! ! Importance sampling works when sampling from proposal is easy, target is hard An alternative scheme… n 1 p(xi ) E p(x)[ f (x)] " # f (xi ) for xi ~ q(x) n i=1 q(xi ) n p(xi -

Hierarchical Models & Bayesian Model Selection
Hierarchical Models & Bayesian Model Selection Geoffrey Roeder Departments of Computer Science and Statistics University of British Columbia Jan. 20, 2016 Geoffrey Roeder Hierarchical Models & Bayesian Model Selection Jan. 20, 2016 Contact information Please report any typos or errors to geoff[email protected] Geoffrey Roeder Hierarchical Models & Bayesian Model Selection Jan. 20, 2016 Outline 1 Hierarchical Bayesian Modelling Coin toss redux: point estimates for θ Hierarchical models Application to clinical study 2 Bayesian Model Selection Introduction Bayes Factors Shortcut for Marginal Likelihood in Conjugate Case Geoffrey Roeder Hierarchical Models & Bayesian Model Selection Jan. 20, 2016 Outline 1 Hierarchical Bayesian Modelling Coin toss redux: point estimates for θ Hierarchical models Application to clinical study 2 Bayesian Model Selection Introduction Bayes Factors Shortcut for Marginal Likelihood in Conjugate Case Geoffrey Roeder Hierarchical Models & Bayesian Model Selection Jan. 20, 2016 Let Y be a random variable denoting number of observed heads in n coin tosses. Then, we can model Y ∼ Bin(n; θ), with probability mass function n p(Y = y j θ) = θy (1 − θ)n−y (1) y We want to estimate the parameter θ Coin toss: point estimates for θ Probability model Consider the experiment of tossing a coin n times. Each toss results in heads with probability θ and tails with probability 1 − θ Geoffrey Roeder Hierarchical Models & Bayesian Model Selection Jan. 20, 2016 We want to estimate the parameter θ Coin toss: point estimates for θ Probability model Consider the experiment of tossing a coin n times. Each toss results in heads with probability θ and tails with probability 1 − θ Let Y be a random variable denoting number of observed heads in n coin tosses.