Bayesian Networks: Inference and Learning
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -

Markov Chain Monte Carlo Convergence Diagnostics: a Comparative Review
Markov Chain Monte Carlo Convergence Diagnostics: A Comparative Review By Mary Kathryn Cowles and Bradley P. Carlin1 Abstract A critical issue for users of Markov Chain Monte Carlo (MCMC) methods in applications is how to determine when it is safe to stop sampling and use the samples to estimate characteristics of the distribu- tion of interest. Research into methods of computing theoretical convergence bounds holds promise for the future but currently has yielded relatively little that is of practical use in applied work. Consequently, most MCMC users address the convergence problem by applying diagnostic tools to the output produced by running their samplers. After giving a brief overview of the area, we provide an expository review of thirteen convergence diagnostics, describing the theoretical basis and practical implementation of each. We then compare their performance in two simple models and conclude that all the methods can fail to detect the sorts of convergence failure they were designed to identify. We thus recommend a combination of strategies aimed at evaluating and accelerating MCMC sampler convergence, including applying diagnostic procedures to a small number of parallel chains, monitoring autocorrelations and cross- correlations, and modifying parameterizations or sampling algorithms appropriately. We emphasize, however, that it is not possible to say with certainty that a finite sample from an MCMC algorithm is rep- resentative of an underlying stationary distribution. 1 Mary Kathryn Cowles is Assistant Professor of Biostatistics, Harvard School of Public Health, Boston, MA 02115. Bradley P. Carlin is Associate Professor, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455. -

Meta-Learning MCMC Proposals
Meta-Learning MCMC Proposals Tongzhou Wang∗ Yi Wu Facebook AI Research University of California, Berkeley [email protected] [email protected] David A. Moorey Stuart J. Russell Google University of California, Berkeley [email protected] [email protected] Abstract Effective implementations of sampling-based probabilistic inference often require manually constructed, model-specific proposals. Inspired by recent progresses in meta-learning for training learning agents that can generalize to unseen environ- ments, we propose a meta-learning approach to building effective and generalizable MCMC proposals. We parametrize the proposal as a neural network to provide fast approximations to block Gibbs conditionals. The learned neural proposals generalize to occurrences of common structural motifs across different models, allowing for the construction of a library of learned inference primitives that can accelerate inference on unseen models with no model-specific training required. We explore several applications including open-universe Gaussian mixture models, in which our learned proposals outperform a hand-tuned sampler, and a real-world named entity recognition task, in which our sampler yields higher final F1 scores than classical single-site Gibbs sampling. 1 Introduction Model-based probabilistic inference is a highly successful paradigm for machine learning, with applications to tasks as diverse as movie recommendation [31], visual scene perception [17], music transcription [3], etc. People learn and plan using mental models, and indeed the entire enterprise of modern science can be viewed as constructing a sophisticated hierarchy of models of physical, mental, and social phenomena. Probabilistic programming provides a formal representation of models as sample-generating programs, promising the ability to explore a even richer range of models. -

Common Quandaries and Their Practical Solutions in Bayesian Network Modeling
Ecological Modelling 358 (2017) 1–9 Contents lists available at ScienceDirect Ecological Modelling journa l homepage: www.elsevier.com/locate/ecolmodel Common quandaries and their practical solutions in Bayesian network modeling Bruce G. Marcot U.S. Forest Service, Pacific Northwest Research Station, 620 S.W. Main Street, Suite 400, Portland, OR, USA a r t i c l e i n f o a b s t r a c t Article history: Use and popularity of Bayesian network (BN) modeling has greatly expanded in recent years, but many Received 21 December 2016 common problems remain. Here, I summarize key problems in BN model construction and interpretation, Received in revised form 12 May 2017 along with suggested practical solutions. Problems in BN model construction include parameterizing Accepted 13 May 2017 probability values, variable definition, complex network structures, latent and confounding variables, Available online 24 May 2017 outlier expert judgments, variable correlation, model peer review, tests of calibration and validation, model overfitting, and modeling wicked problems. Problems in BN model interpretation include objec- Keywords: tive creep, misconstruing variable influence, conflating correlation with causation, conflating proportion Bayesian networks and expectation with probability, and using expert opinion. Solutions are offered for each problem and Modeling problems researchers are urged to innovate and share further solutions. Modeling solutions Bias Published by Elsevier B.V. Machine learning Expert knowledge 1. Introduction other fields. Although the number of generally accessible journal articles on BN modeling has continued to increase in recent years, Bayesian network (BN) models are essentially graphs of vari- achieving an exponential growth at least during the period from ables depicted and linked by probabilities (Koski and Noble, 2011). -
3.3 Bayes' Formula
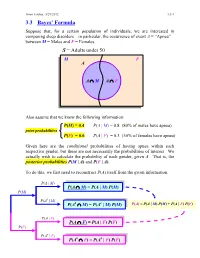
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Markov Chain Monte Carlo Edps 590BAY
Markov Chain Monte Carlo Edps 590BAY Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Fall 2019 Overivew Bayesian computing Markov Chains Metropolis Example Practice Assessment Metropolis 2 Practice Overview ◮ Introduction to Bayesian computing. ◮ Markov Chain ◮ Metropolis algorithm for mean of normal given fixed variance. ◮ Revisit anorexia data. ◮ Practice problem with SAT data. ◮ Some tools for assessing convergence ◮ Metropolis algorithm for mean and variance of normal. ◮ Anorexia data. ◮ Practice problem. ◮ Summary Depending on the book that you select for this course, read either Gelman et al. pp 2751-291 or Kruschke Chapters pp 143–218. I am relying more on Gelman et al. C.J. Anderson (Illinois) MCMC Fall 2019 2.2/ 45 Overivew Bayesian computing Markov Chains Metropolis Example Practice Assessment Metropolis 2 Practice Introduction to Bayesian Computing ◮ Our major goal is to approximate the posterior distributions of unknown parameters and use them to estimate parameters. ◮ The analytic computations are fine for simple problems, ◮ Beta-binomial for bounded counts ◮ Normal-normal for continuous variables ◮ Gamma-Poisson for (unbounded) counts ◮ Dirichelt-Multinomial for multicategory variables (i.e., a categorical variable) ◮ Models in the exponential family with small number of parameters ◮ For large number of parameters and more complex models ◮ Algebra of analytic solution becomes overwhelming ◮ Grid takes too much time. ◮ Too difficult for most applications. C.J. Anderson (Illinois) MCMC Fall 2019 3.3/ 45 Overivew Bayesian computing Markov Chains Metropolis Example Practice Assessment Metropolis 2 Practice Steps in Modeling Recall that the steps in an analysis: 1. Choose model for data (i.e., p(y θ)) and model for parameters | (i.e., p(θ) and p(θ y)). -

Bayesian Network Modelling with Examples
Bayesian Network Modelling with Examples Marco Scutari [email protected] Department of Statistics University of Oxford November 28, 2016 What Are Bayesian Networks? Marco Scutari University of Oxford What Are Bayesian Networks? A Graph and a Probability Distribution Bayesian networks (BNs) are defined by: a network structure, a directed acyclic graph G = (V;A), in which each node vi 2 V corresponds to a random variable Xi; a global probability distribution X with parameters Θ, which can be factorised into smaller local probability distributions according to the arcs aij 2 A present in the graph. The main role of the network structure is to express the conditional independence relationships among the variables in the model through graphical separation, thus specifying the factorisation of the global distribution: p Y P(X) = P(Xi j ΠXi ;ΘXi ) where ΠXi = fparents of Xig i=1 Marco Scutari University of Oxford What Are Bayesian Networks? How the DAG Maps to the Probability Distribution Graphical Probabilistic DAG separation independence A B C E D F Formally, the DAG is an independence map of the probability distribution of X, with graphical separation (??G) implying probabilistic independence (??P ). Marco Scutari University of Oxford What Are Bayesian Networks? Graphical Separation in DAGs (Fundamental Connections) separation (undirected graphs) A B C d-separation (directed acyclic graphs) A B C A B C A B C Marco Scutari University of Oxford What Are Bayesian Networks? Graphical Separation in DAGs (General Case) Now, in the general case we can extend the patterns from the fundamental connections and apply them to every possible path between A and B for a given C; this is how d-separation is defined. -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 . -

The Bayesian Approach to Statistics
THE BAYESIAN APPROACH TO STATISTICS ANTHONY O’HAGAN INTRODUCTION the true nature of scientific reasoning. The fi- nal section addresses various features of modern By far the most widely taught and used statisti- Bayesian methods that provide some explanation for the rapid increase in their adoption since the cal methods in practice are those of the frequen- 1980s. tist school. The ideas of frequentist inference, as set out in Chapter 5 of this book, rest on the frequency definition of probability (Chapter 2), BAYESIAN INFERENCE and were developed in the first half of the 20th century. This chapter concerns a radically differ- We first present the basic procedures of Bayesian ent approach to statistics, the Bayesian approach, inference. which depends instead on the subjective defini- tion of probability (Chapter 3). In some respects, Bayesian methods are older than frequentist ones, Bayes’s Theorem and the Nature of Learning having been the basis of very early statistical rea- Bayesian inference is a process of learning soning as far back as the 18th century. Bayesian from data. To give substance to this statement, statistics as it is now understood, however, dates we need to identify who is doing the learning and back to the 1950s, with subsequent development what they are learning about. in the second half of the 20th century. Over that time, the Bayesian approach has steadily gained Terms and Notation ground, and is now recognized as a legitimate al- ternative to the frequentist approach. The person doing the learning is an individual This chapter is organized into three sections. -

Bayesian Phylogenetics and Markov Chain Monte Carlo Will Freyman
IB200, Spring 2016 University of California, Berkeley Lecture 12: Bayesian phylogenetics and Markov chain Monte Carlo Will Freyman 1 Basic Probability Theory Probability is a quantitative measurement of the likelihood of an outcome of some random process. The probability of an event, like flipping a coin and getting heads is notated P (heads). The probability of getting tails is then 1 − P (heads) = P (tails). • The joint probability of event A and event B both occuring is written as P (A; B). The joint probability of mutually exclusive events, like flipping a coin once and getting heads and tails, is 0. • The probability of A occuring given that B has already occurred is written P (AjB), which is read \the probability of A given B". This is a conditional probability. • The marginal probability of A is P (A), which is calculated by summing or integrating the joint probability over B. In other words, P (A) = P (A; B) + P (A; not B). • Joint, conditional, and marginal probabilities can be combined with the expression P (A; B) = P (A)P (BjA) = P (B)P (AjB). • The above expression can be rearranged into Bayes' theorem: P (BjA)P (A) P (AjB) = P (B) Bayes' theorem is a straightforward and uncontroversial way to calculate an inverse conditional probability. • If P (AjB) = P (A) and P (BjA) = P (B) then A and B are independent. Then the joint probability is calculated P (A; B) = P (A)P (B). 2 Interpretations of Probability What exactly is a probability? Does it really exist? There are two major interpretations of probability: • Frequentists believe that the probability of an event is its relative frequency over time. -

Empirical Evaluation of Scoring Functions for Bayesian Network Model Selection
Liu et al. BMC Bioinformatics 2012, 13(Suppl 15):S14 http://www.biomedcentral.com/1471-2105/13/S15/S14 PROCEEDINGS Open Access Empirical evaluation of scoring functions for Bayesian network model selection Zhifa Liu1,2†, Brandon Malone1,3†, Changhe Yuan1,4* From Proceedings of the Ninth Annual MCBIOS Conference. Dealing with the Omics Data Deluge Oxford, MS, USA. 17-18 February 2012 Abstract In this work, we empirically evaluate the capability of various scoring functions of Bayesian networks for recovering true underlying structures. Similar investigations have been carried out before, but they typically relied on approximate learning algorithms to learn the network structures. The suboptimal structures found by the approximation methods have unknown quality and may affect the reliability of their conclusions. Our study uses an optimal algorithm to learn Bayesian network structures from datasets generated from a set of gold standard Bayesian networks. Because all optimal algorithms always learn equivalent networks, this ensures that only the choice of scoring function affects the learned networks. Another shortcoming of the previous studies stems from their use of random synthetic networks as test cases. There is no guarantee that these networks reflect real-world data. We use real-world data to generate our gold-standard structures, so our experimental design more closely approximates real-world situations. A major finding of our study suggests that, in contrast to results reported by several prior works, the Minimum Description Length (MDL) (or equivalently, Bayesian information criterion (BIC)) consistently outperforms other scoring functions such as Akaike’s information criterion (AIC), Bayesian Dirichlet equivalence score (BDeu), and factorized normalized maximum likelihood (fNML) in recovering the underlying Bayesian network structures. -

Paradoxes and Priors in Bayesian Regression
Paradoxes and Priors in Bayesian Regression Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Agniva Som, B. Stat., M. Stat. Graduate Program in Statistics The Ohio State University 2014 Dissertation Committee: Dr. Christopher M. Hans, Advisor Dr. Steven N. MacEachern, Co-advisor Dr. Mario Peruggia c Copyright by Agniva Som 2014 Abstract The linear model has been by far the most popular and most attractive choice of a statistical model over the past century, ubiquitous in both frequentist and Bayesian literature. The basic model has been gradually improved over the years to deal with stronger features in the data like multicollinearity, non-linear or functional data pat- terns, violation of underlying model assumptions etc. One valuable direction pursued in the enrichment of the linear model is the use of Bayesian methods, which blend information from the data likelihood and suitable prior distributions placed on the unknown model parameters to carry out inference. This dissertation studies the modeling implications of many common prior distri- butions in linear regression, including the popular g prior and its recent ameliorations. Formalization of desirable characteristics for model comparison and parameter esti- mation has led to the growth of appropriate mixtures of g priors that conform to the seven standard model selection criteria laid out by Bayarri et al. (2012). The existence of some of these properties (or lack thereof) is demonstrated by examining the behavior of the prior under suitable limits on the likelihood or on the prior itself.