Phyre 2 Results for P15288
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases
International Journal of Molecular Sciences Review Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases Peter Goettig Structural Biology Group, Faculty of Molecular Biology, University of Salzburg, Billrothstrasse 11, 5020 Salzburg, Austria; [email protected]; Tel.: +43-662-8044-7283; Fax: +43-662-8044-7209 Academic Editor: Cheorl-Ho Kim Received: 30 July 2016; Accepted: 10 November 2016; Published: 25 November 2016 Abstract: Posttranslational modifications are an important feature of most proteases in higher organisms, such as the conversion of inactive zymogens into active proteases. To date, little information is available on the role of glycosylation and functional implications for secreted proteases. Besides a stabilizing effect and protection against proteolysis, several proteases show a significant influence of glycosylation on the catalytic activity. Glycans can alter the substrate recognition, the specificity and binding affinity, as well as the turnover rates. However, there is currently no known general pattern, since glycosylation can have both stimulating and inhibiting effects on activity. Thus, a comparative analysis of individual cases with sufficient enzyme kinetic and structural data is a first approach to describe mechanistic principles that govern the effects of glycosylation on the function of proteases. The understanding of glycan functions becomes highly significant in proteomic and glycomic studies, which demonstrated that cancer-associated proteases, such as kallikrein-related peptidase 3, exhibit strongly altered glycosylation patterns in pathological cases. Such findings can contribute to a variety of future biomedical applications. Keywords: secreted protease; sequon; N-glycosylation; O-glycosylation; core glycan; enzyme kinetics; substrate recognition; flexible loops; Michaelis constant; turnover number 1. -

JASON MARC GOLDSTEIN the Isolation, Characterization
JASON MARC GOLDSTEIN The Isolation, Characterization and Cloning of Three Novel Peptidases From Streptoccocus gordonii: Their Potential Roles in Subacute Bacterial Endocarditis (Under the Direction of JAMES TRAVIS) Streptococcus gordonii is generally considered a benign inhabitant of the oral microflora yet is a primary etiological agent in the development of subacute bacterial endocarditis (SBE), an inflammatory state that propagates thrombus formation and tissue damage on the surface of heart valves. Colonization and adherence mechanisms have been identified, yet factors necessary to sustain growth remain unidentified. Strain FSS2 produced three extracellular aminopeptidase activities during growth in neutral pH- controlled batch cultures. The first included a serine-class dipeptidyl-aminopeptidase, an x-Pro DPP (Sg-xPDPP) found as an 85 kDa monomer by SDS-PAGE while appearing as a homodimer under native conditions. Kinetic studies indicated a unique and stringent x- Pro specificity comparable to the DPPIV/CD26 and lactococcal x-Pro DPP families. Isolation of the full-length gene uncovered a 759-amino acid polypeptide with a mass of 87,115 Da and theoretical pI of 5.6. Significant homology was found with PepX gene family members from Lactobacillus ssp. and Lactococcus ssp., and putative streptococcal x-Pro DPPs. The second activity was a putative serine-class arginine aminopeptidase (Sg- RAP) with some cysteine-class characteristics. It was found as a protein monomer of 70 kDa under denaturing conditions. Nested PCR cloning enabled the isolation of a 324 bp- long DNA fragment encoding the protein’s 108 amino acid N-terminus. Culture activity profiles and N-terminal sequence analysis indicated the release of this protein from the cell surface. -

University of California, San Diego
UNIVERSITY OF CALIFORNIA, SAN DIEGO A Lipidomic Perspective on Inflammatory Macrophage Eicosanoid Signaling A Thesis submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry by Paul Christopher Norris Committee in charge: Professor Edward A. Dennis, Chair Professor Pieter C. Dorrestein Professor Partho Ghosh Professor Christopher K. Glass Professor Michael J. Sailor 2013 The Dissertation of Paul Christopher Norris is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Chair University of California, San Diego 2013 iii DEDICATION To my parents, Darrell and Kathy, for always allowing me to think (and choose) for myself. iv TABLE OF CONTENTS Signature page ............................................................................................................................ iii Dedication .................................................................................................................................. iv Table of contents ......................................................................................................................... v List of symbols and abbreviations ........................................................................................... viii List of figures ............................................................................................................................. xi List of tables ............................................................................................................................ -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -

MALE Protein Name Accession Number Molecular Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean H Mean PDAC Mean T-Test PDAC Vs. H T-Test
MALE t-test t-test Accession Molecular H PDAC PDAC vs. PDAC vs. Protein Name Number Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean Mean Mean H CP PDAC/H PDAC/CP - 22 kDa protein IPI00219910 22 kDa 7 5 4 8 1 0 6 6 1 0.1126 0.0456 0.1 0.1 - Cold agglutinin FS-1 L-chain (Fragment) IPI00827773 12 kDa 32 39 34 26 53 57 36 30 55 0.0309 0.0388 1.8 1.5 - HRV Fab 027-VL (Fragment) IPI00827643 12 kDa 4 6 0 0 0 0 5 0 0 - 0.0574 - 0.0 - REV25-2 (Fragment) IPI00816794 15 kDa 8 12 5 7 8 9 10 6 8 0.2225 0.3844 1.3 0.8 A1BG Alpha-1B-glycoprotein precursor IPI00022895 54 kDa 115 109 106 112 111 100 112 109 105 0.6497 0.4138 1.0 0.9 A2M Alpha-2-macroglobulin precursor IPI00478003 163 kDa 62 63 86 72 14 18 63 79 16 0.0120 0.0019 0.2 0.3 ABCB1 Multidrug resistance protein 1 IPI00027481 141 kDa 41 46 23 26 52 64 43 25 58 0.0355 0.1660 2.4 1.3 ABHD14B Isoform 1 of Abhydrolase domain-containing proteinIPI00063827 14B 22 kDa 19 15 19 17 15 9 17 18 12 0.2502 0.3306 0.7 0.7 ABP1 Isoform 1 of Amiloride-sensitive amine oxidase [copper-containing]IPI00020982 precursor85 kDa 1 5 8 8 0 0 3 8 0 0.0001 0.2445 0.0 0.0 ACAN aggrecan isoform 2 precursor IPI00027377 250 kDa 38 30 17 28 34 24 34 22 29 0.4877 0.5109 1.3 0.8 ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somaticIPI00437751 isoform precursor150 kDa 48 34 67 56 28 38 41 61 33 0.0600 0.4301 0.5 0.8 ACE2 Isoform 1 of Angiotensin-converting enzyme 2 precursorIPI00465187 92 kDa 11 16 20 30 4 5 13 25 5 0.0557 0.0847 0.2 0.4 ACO1 Cytoplasmic aconitate hydratase IPI00008485 98 kDa 2 2 0 0 0 0 2 0 0 - 0.0081 - 0.0 -
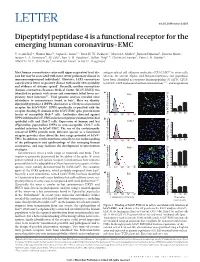
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells. -

Novel Proteins Regulated by Mtor in Subependymal Giant Cell Astrocytomas of Patients with Tuberous Sclerosis Complex and New Therapeutic Implications
The American Journal of Pathology, Vol. 176, No. 4, April 2010 Copyright © American Society for Investigative Pathology DOI: 10.2353/ajpath.2010.090950 Molecular Pathogenesis of Genetic and Inherited Diseases Novel Proteins Regulated by mTOR in Subependymal Giant Cell Astrocytomas of Patients with Tuberous Sclerosis Complex and New Therapeutic Implications Magdalena Ewa Tyburczy,* Katarzyna Kotulska,† and demonstrated an effective modulation of SEGA Piotr Pokarowski,‡ Jakub Mieczkowski,* growth by pharmacological inhibition of both Joanna Kucharska,* Wieslawa Grajkowska,† mTOR and extracellular signal-regulated kinase sig- Maciej Roszkowski,§ Sergiusz Jozwiak,† naling pathways, which could represent a novel ther- and Bozena Kaminska* apeutic approach. (Am J Pathol 2010, 176:1878–1890; DOI: 10.2353/ajpath.2010.090950) From the Laboratory of Transcription Regulation,* the Nencki Institute of Experimental Biology, Warsaw; the Departments of Neurology and Epileptology,† and Neurosurgery,§ the Children’s Subependymal giant cell astrocytomas (SEGAs) are rare, Memorial Health Institute, Warsaw; and the Faculty of low-grade brain tumors (World Health Organization Grade I) Mathematics, Informatics, and Mechanics,‡ University of of a mixed glioneuronal lineage.1,2 They are observed in Warsaw, Poland 10% to 20% of patients with tuberous sclerosis complex (TSC) and are the major cause of morbidity in children and young adults with TSC.3 The disease affects about one in Subependymal giant cell astrocytomas (SEGAs) are 6000 people, is characterized by the formation of benign rare brain tumors associated with tuberous sclerosis tumors in multiple organs (mainly brain, heart, kidneys, skin, complex (TSC), a disease caused by mutations in TSC1 or lungs), and is often associated with epilepsy, mental or TSC2, resulting in enhancement of mammalian retardation, and autism.4,5 Tuberous sclerosis complex is target of rapamycin (mTOR) activity, dysregulation of caused by mutation in one of two tumor suppressor genes, cell growth, and tumorigenesis. -

And Exopeptidases in a Processing Enzyme System: Activation
Proc. Nail. Acad. Sci. USA Vol. 85, pp. 5468-5472, August 1988 Biochemistry Relationship between endo- and exopeptidases in a processing enzyme system: Activation of an endoprotease by the aminopeptidase B-like activity in somatostatin-28 convertase (brain cortex/basic amino acid pairs/peptide substrates/protease inhibitors/prohormone maturation) SOPHIE GOMEZ, PABLO GLUSCHANKOF, AGNES LEPAGE, AND PAUL COHEN Groupe de Neurobiochimie Cellulaire et Moldculaire, Universitd Pierre et Marie Curie, Unitd Associde 554 au Centre National de la Recherche Scientifique, 96 boulevard Raspail, 75006 Paris, France Communicated by I. Robert Lehman, April 8, 1988 (receivedfor review December 15, 1987) ABSTRACT The somatostatin-28 convertase activity in- somatostatin-14 and the amino-terminal dodecapeptide so- volved in vitro in the processing of somatostatin-28 into the matostatin-28-(1-12) (16). neuropeptides somatostatin-28-(1-12) and somatostatin-14 is We have described an endoprotease that cleaves the composed of an endoprotease and a basic aminopeptidase. We peptide bond on the amino side ofthe Arg-Lys doublet in the report herein on the purification to apparent homogeneity of somatostatin-28 sequence (17, 18), releasing the somato- these two constituents and on their functional interrelationship. statin-28-(1-12) fragment and [Arg-2,Lys-1]somatostatin-14. In particular we observed that after various physicochemical The released [Arg-2,Lys-']somatostatin-14 is further proc- treatments, the 90-kDa endoprotease activity was recovered essed by an aminopeptidase B-like activity (18, 19) that is also both at this molecular mass and as a 45-kDa entity. Moreover, present in the preparation. We report herein the purification the production of [Arg2,LysJllsomatostatin-14 from somato- to apparent homogeneity ofthese two activities. -

Recombinant Human Dipeptidase 1 Protein
Leader in Biomolecular Solutions for Life Science Recombinant Human Dipeptidase 1 Protein Catalog No.: RP01097 Recombinant Sequence Information Background Species Gene ID Swiss Prot This protein is a kidney membrane enzyme involved in the metabolism of Human 1800 P16444 glutathione and other similar proteins by dipeptide hydrolysis. The encoded protein is known to regulate leukotriene activity by catalyzing the conversion of Tags leukotriene D4 to leukotriene E4. This protein uses zinc as a cofactor and acts as a C-6×His disulfide-linked homodimer. Two transcript variants encoding the same protein have been found for this gene. Synonyms Dipeptidase 1; Dehydropeptidase-I; Microsomal Dipeptidase; Renal Basic Information Dipeptidase; hRDP; DPEP1; MDP; RDP Description Recombinant Human Dipeptidase 1 Protein is produced by Mammalian expression Product Information system. The target protein is expressed with sequence (Asp17-Ser385) of human Dipeptidase 1 (Accession #P16444) fused with a 6×His tag at the C-terminus. Source Purification Mammalian > 95% by SDS- Bio-Activity PAGE. Storage Endotoxin Store the lyophilized protein at -20°C to -80 °C for long term. < 1.0 EU/μg of the protein by LAL After reconstitution, the protein solution is stable at -20 °C for 3 months, at 2-8 °C method. for up to 1 week. Avoid repeated freeze/thaw cycles. Formulation Lyophilized from a 0.22 μm filtered solution of 20mM PB, 150mM NaCl, pH7.4.Contact us for customized product form or formulation. Reconstitution Reconstitute to a concentration of 0.1-0.5 mg/mL in sterile distilled water. Contact www.abclonal.com Validation Data Recombinant Human Dipeptidase 1 Protein was determined by SDS-PAGE with Coomassie Blue, showing a band at 41 kDa. -

Proteolytic Cleavage—Mechanisms, Function
Review Cite This: Chem. Rev. 2018, 118, 1137−1168 pubs.acs.org/CR Proteolytic CleavageMechanisms, Function, and “Omic” Approaches for a Near-Ubiquitous Posttranslational Modification Theo Klein,†,⊥ Ulrich Eckhard,†,§ Antoine Dufour,†,¶ Nestor Solis,† and Christopher M. Overall*,†,‡ † ‡ Life Sciences Institute, Department of Oral Biological and Medical Sciences, and Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada ABSTRACT: Proteases enzymatically hydrolyze peptide bonds in substrate proteins, resulting in a widespread, irreversible posttranslational modification of the protein’s structure and biological function. Often regarded as a mere degradative mechanism in destruction of proteins or turnover in maintaining physiological homeostasis, recent research in the field of degradomics has led to the recognition of two main yet unexpected concepts. First, that targeted, limited proteolytic cleavage events by a wide repertoire of proteases are pivotal regulators of most, if not all, physiological and pathological processes. Second, an unexpected in vivo abundance of stable cleaved proteins revealed pervasive, functionally relevant protein processing in normal and diseased tissuefrom 40 to 70% of proteins also occur in vivo as distinct stable proteoforms with undocumented N- or C- termini, meaning these proteoforms are stable functional cleavage products, most with unknown functional implications. In this Review, we discuss the structural biology aspects and mechanisms -

Contribution of Endo- and Exopeptidases to Formation of Non-Protein N During Ensiling of Alfalfa
Contribution of endo- and exopeptidases to formation of non-protein N during ensiling of alfalfa Dr. Xusheng Guo,1 W. Cheng,1 L. Tao,2 Yu Zhu,2 F.Y. Yang 2 & H. Zhou2 1.School of Life Science, Lanzhou University 1. International Centre for Tibetan Plateau Ecosystem Management (ICTPEM), Lanzhou University 2. Institute of Grassland Science, China Agricultural University Hämeenlinna, Finland 2-4 July 2012 Introduction Alfalfa (Medicago Sativa L.) is well known for its high nutritive value However, after ensiling: N use efficiency Extensive ppyroteolysis Reduce True protein NPN(Peptide, FAA, NH3-N etc.) Silage DM intake (44-87% of Total N; Muck, 1987) Silage Fermentation Proteolysis in ensiled forage mainly results from plant proteases (Ohshima and McDonald, 1978; McKersie, 1981; Heron et al., 1988). Proteases (peptidases) are divided into 2 classes (NC-IUBMB, 1992): Exopeptidase Endopeptidase Objectives Proteases (peptidases, E.C.3.4) Endopeptidases Exopeptidases ?! Ser ine pep tidase (E .C .3 .4 .21) Aminopeptidase (EC 3.4.11) Carboxypeptidase (EC 3.4.16) Cysteine peptidase (E.C.3.4.22) Dipeptidase (EC 3.4.13) Aspartic Peptidase (E.C.3.4.23) Dipeptidyl-peptidase (EC 3.4.14) Metallopeptidase (E.C.3.4.24) Tripeptidyl-peptidase (EC 3.4.14) Peptidyl-dipeptidase (EC 3.4.15) Aims of our research were: 1. To clarify the classes of exo- and endopeptidases that are involved in proteolysis within ensiled alfalfa. 2. To determine the contribution of these peptidases to the formation of different NPN compounds (peptide-N, FAA-N, and NH3-N) during -

Thematic Review
Supplemental Material can be found at: http://www.jlr.org/content/suppl/2009/09/16/R900004-JLR20 0.DC1.html thematic review Thematic Review Series: Proteomics An integrated omics analysis of eicosanoid biology1 Matthew W. Buczynski, Darren S. Dumlao, and Edward A. Dennis2 Department of Chemistry and Biochemistry, Department of Pharmacology, and School of Medicine, University of California, San Diego, La Jolla, CA 92093 Abstract Eicosanoids have been implicated in a vast number to address the question of how molecular biology works as of devastating inflammatory conditions, including arthritis, an integrated process (1). atherosclerosis, pain, and cancer. Currently, over a hundred Systems biology has advanced exponentially during the different eicosanoids have been identified, with many having past two decades, with transcriptomics, proteomics, and potent bioactive signaling capacity. These lipid metabolites metabolomics each playing an integral role. Each of these are synthesized de novo by at least 50 unique enzymes, many of which have been cloned and characterized. Due to the ex- platforms brings its own unique advantages and limitations Downloaded from tensive characterization of eicosanoid biosynthetic pathways, in facilitating the investigation of disease pathology. A this field provides a unique framework for integrating geno- transcriptomic approach can detect the upregulation and mics, proteomics, and metabolomics toward the investigation downregulation of important biosynthetic and signaling of disease pathology. To facilitate a concerted systems biol- genes; however, gene changes often donʼt directly corre- ogy approach, this review outlines the proteins implicated in late with changes in protein levels (2). Proteomic analy- eicosanoid biosynthesis and signaling in human, mouse, and ses can identify enzymes and posttranslational protein www.jlr.org rat.