Thematic Review
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases
International Journal of Molecular Sciences Review Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases Peter Goettig Structural Biology Group, Faculty of Molecular Biology, University of Salzburg, Billrothstrasse 11, 5020 Salzburg, Austria; [email protected]; Tel.: +43-662-8044-7283; Fax: +43-662-8044-7209 Academic Editor: Cheorl-Ho Kim Received: 30 July 2016; Accepted: 10 November 2016; Published: 25 November 2016 Abstract: Posttranslational modifications are an important feature of most proteases in higher organisms, such as the conversion of inactive zymogens into active proteases. To date, little information is available on the role of glycosylation and functional implications for secreted proteases. Besides a stabilizing effect and protection against proteolysis, several proteases show a significant influence of glycosylation on the catalytic activity. Glycans can alter the substrate recognition, the specificity and binding affinity, as well as the turnover rates. However, there is currently no known general pattern, since glycosylation can have both stimulating and inhibiting effects on activity. Thus, a comparative analysis of individual cases with sufficient enzyme kinetic and structural data is a first approach to describe mechanistic principles that govern the effects of glycosylation on the function of proteases. The understanding of glycan functions becomes highly significant in proteomic and glycomic studies, which demonstrated that cancer-associated proteases, such as kallikrein-related peptidase 3, exhibit strongly altered glycosylation patterns in pathological cases. Such findings can contribute to a variety of future biomedical applications. Keywords: secreted protease; sequon; N-glycosylation; O-glycosylation; core glycan; enzyme kinetics; substrate recognition; flexible loops; Michaelis constant; turnover number 1. -

University of California, San Diego
UNIVERSITY OF CALIFORNIA, SAN DIEGO A Lipidomic Perspective on Inflammatory Macrophage Eicosanoid Signaling A Thesis submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry by Paul Christopher Norris Committee in charge: Professor Edward A. Dennis, Chair Professor Pieter C. Dorrestein Professor Partho Ghosh Professor Christopher K. Glass Professor Michael J. Sailor 2013 The Dissertation of Paul Christopher Norris is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Chair University of California, San Diego 2013 iii DEDICATION To my parents, Darrell and Kathy, for always allowing me to think (and choose) for myself. iv TABLE OF CONTENTS Signature page ............................................................................................................................ iii Dedication .................................................................................................................................. iv Table of contents ......................................................................................................................... v List of symbols and abbreviations ........................................................................................... viii List of figures ............................................................................................................................. xi List of tables ............................................................................................................................ -

MALE Protein Name Accession Number Molecular Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean H Mean PDAC Mean T-Test PDAC Vs. H T-Test
MALE t-test t-test Accession Molecular H PDAC PDAC vs. PDAC vs. Protein Name Number Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean Mean Mean H CP PDAC/H PDAC/CP - 22 kDa protein IPI00219910 22 kDa 7 5 4 8 1 0 6 6 1 0.1126 0.0456 0.1 0.1 - Cold agglutinin FS-1 L-chain (Fragment) IPI00827773 12 kDa 32 39 34 26 53 57 36 30 55 0.0309 0.0388 1.8 1.5 - HRV Fab 027-VL (Fragment) IPI00827643 12 kDa 4 6 0 0 0 0 5 0 0 - 0.0574 - 0.0 - REV25-2 (Fragment) IPI00816794 15 kDa 8 12 5 7 8 9 10 6 8 0.2225 0.3844 1.3 0.8 A1BG Alpha-1B-glycoprotein precursor IPI00022895 54 kDa 115 109 106 112 111 100 112 109 105 0.6497 0.4138 1.0 0.9 A2M Alpha-2-macroglobulin precursor IPI00478003 163 kDa 62 63 86 72 14 18 63 79 16 0.0120 0.0019 0.2 0.3 ABCB1 Multidrug resistance protein 1 IPI00027481 141 kDa 41 46 23 26 52 64 43 25 58 0.0355 0.1660 2.4 1.3 ABHD14B Isoform 1 of Abhydrolase domain-containing proteinIPI00063827 14B 22 kDa 19 15 19 17 15 9 17 18 12 0.2502 0.3306 0.7 0.7 ABP1 Isoform 1 of Amiloride-sensitive amine oxidase [copper-containing]IPI00020982 precursor85 kDa 1 5 8 8 0 0 3 8 0 0.0001 0.2445 0.0 0.0 ACAN aggrecan isoform 2 precursor IPI00027377 250 kDa 38 30 17 28 34 24 34 22 29 0.4877 0.5109 1.3 0.8 ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somaticIPI00437751 isoform precursor150 kDa 48 34 67 56 28 38 41 61 33 0.0600 0.4301 0.5 0.8 ACE2 Isoform 1 of Angiotensin-converting enzyme 2 precursorIPI00465187 92 kDa 11 16 20 30 4 5 13 25 5 0.0557 0.0847 0.2 0.4 ACO1 Cytoplasmic aconitate hydratase IPI00008485 98 kDa 2 2 0 0 0 0 2 0 0 - 0.0081 - 0.0 -

Novel Proteins Regulated by Mtor in Subependymal Giant Cell Astrocytomas of Patients with Tuberous Sclerosis Complex and New Therapeutic Implications
The American Journal of Pathology, Vol. 176, No. 4, April 2010 Copyright © American Society for Investigative Pathology DOI: 10.2353/ajpath.2010.090950 Molecular Pathogenesis of Genetic and Inherited Diseases Novel Proteins Regulated by mTOR in Subependymal Giant Cell Astrocytomas of Patients with Tuberous Sclerosis Complex and New Therapeutic Implications Magdalena Ewa Tyburczy,* Katarzyna Kotulska,† and demonstrated an effective modulation of SEGA Piotr Pokarowski,‡ Jakub Mieczkowski,* growth by pharmacological inhibition of both Joanna Kucharska,* Wieslawa Grajkowska,† mTOR and extracellular signal-regulated kinase sig- Maciej Roszkowski,§ Sergiusz Jozwiak,† naling pathways, which could represent a novel ther- and Bozena Kaminska* apeutic approach. (Am J Pathol 2010, 176:1878–1890; DOI: 10.2353/ajpath.2010.090950) From the Laboratory of Transcription Regulation,* the Nencki Institute of Experimental Biology, Warsaw; the Departments of Neurology and Epileptology,† and Neurosurgery,§ the Children’s Subependymal giant cell astrocytomas (SEGAs) are rare, Memorial Health Institute, Warsaw; and the Faculty of low-grade brain tumors (World Health Organization Grade I) Mathematics, Informatics, and Mechanics,‡ University of of a mixed glioneuronal lineage.1,2 They are observed in Warsaw, Poland 10% to 20% of patients with tuberous sclerosis complex (TSC) and are the major cause of morbidity in children and young adults with TSC.3 The disease affects about one in Subependymal giant cell astrocytomas (SEGAs) are 6000 people, is characterized by the formation of benign rare brain tumors associated with tuberous sclerosis tumors in multiple organs (mainly brain, heart, kidneys, skin, complex (TSC), a disease caused by mutations in TSC1 or lungs), and is often associated with epilepsy, mental or TSC2, resulting in enhancement of mammalian retardation, and autism.4,5 Tuberous sclerosis complex is target of rapamycin (mTOR) activity, dysregulation of caused by mutation in one of two tumor suppressor genes, cell growth, and tumorigenesis. -

Recombinant Human Dipeptidase 1 Protein
Leader in Biomolecular Solutions for Life Science Recombinant Human Dipeptidase 1 Protein Catalog No.: RP01097 Recombinant Sequence Information Background Species Gene ID Swiss Prot This protein is a kidney membrane enzyme involved in the metabolism of Human 1800 P16444 glutathione and other similar proteins by dipeptide hydrolysis. The encoded protein is known to regulate leukotriene activity by catalyzing the conversion of Tags leukotriene D4 to leukotriene E4. This protein uses zinc as a cofactor and acts as a C-6×His disulfide-linked homodimer. Two transcript variants encoding the same protein have been found for this gene. Synonyms Dipeptidase 1; Dehydropeptidase-I; Microsomal Dipeptidase; Renal Basic Information Dipeptidase; hRDP; DPEP1; MDP; RDP Description Recombinant Human Dipeptidase 1 Protein is produced by Mammalian expression Product Information system. The target protein is expressed with sequence (Asp17-Ser385) of human Dipeptidase 1 (Accession #P16444) fused with a 6×His tag at the C-terminus. Source Purification Mammalian > 95% by SDS- Bio-Activity PAGE. Storage Endotoxin Store the lyophilized protein at -20°C to -80 °C for long term. < 1.0 EU/μg of the protein by LAL After reconstitution, the protein solution is stable at -20 °C for 3 months, at 2-8 °C method. for up to 1 week. Avoid repeated freeze/thaw cycles. Formulation Lyophilized from a 0.22 μm filtered solution of 20mM PB, 150mM NaCl, pH7.4.Contact us for customized product form or formulation. Reconstitution Reconstitute to a concentration of 0.1-0.5 mg/mL in sterile distilled water. Contact www.abclonal.com Validation Data Recombinant Human Dipeptidase 1 Protein was determined by SDS-PAGE with Coomassie Blue, showing a band at 41 kDa. -
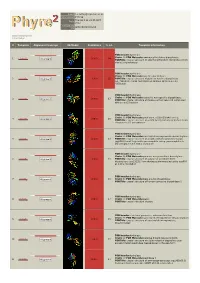
Phyre 2 Results for P15288
Email [email protected] Description P15288 Thu Jan 5 11:34:45 GMT Date 2012 Unique Job d2087d8d303c52c9 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:hydrolase Chain: B: PDB Molecule:aminoacyl-histidine dipeptidase; 1 c3mruB_ 100.0 64 Alignment PDBTitle: crystal structure of aminoacylhistidine dipeptidase from vibrio2 alginolyticus PDB header:hydrolase Chain: B: PDB Molecule:xaa-his dipeptidase; 2 c2qyvB_ Alignment 100.0 55 PDBTitle: crystal structure of putative xaa-his dipeptidase (yp_718209.1) from2 haemophilus somnus 129pt at 2.11 a resolution PDB header:hydrolase Chain: A: PDB Molecule:cytosolic non-specific dipeptidase; 3 c2zogA_ 100.0 17 Alignment PDBTitle: crystal structure of mouse carnosinase cn2 complexed with zn and2 bestatin PDB header:hydrolase Chain: B: PDB Molecule:peptidase, m20/m25/m40 family; 4 c2pokB_ 100.0 16 Alignment PDBTitle: crystal structure of a m20 family metallo peptidase from streptococcus2 pneumoniae PDB header:hydrolase Chain: A: PDB Molecule:succinyl-diaminopimelate desuccinylase; 5 c3pfeA_ Alignment 100.0 13 PDBTitle: crystal structure of a m20a metallo peptidase (dape, lpg0809) from2 legionella pneumophila subsp. pneumophila str. philadelphia 1 at 1.503 a resolution PDB header:hydrolase Chain: B: PDB Molecule:putative acetylornithine deacetylase; 6 c3pfoB_ Alignment 100.0 16 PDBTitle: crystal structure of a putative acetylornithine deacetylase (rpa2325)2 from rhodopseudomonas palustris cga009 at 1.90 a resolution PDB -

Supplementary Table 6 . Pharmacophore Candidates
Supplementary Table 6. pharmacophore candidates identified by pharmMapper Pharma Model Norm Fit symple Name Uniplot 2p3g_v 0.9707 MAPKAPK2 MAP kinase-activated protein kinase 2 P49137 3gam_v 0.8849 NQO2 Ribosyldihydronicotinamide dehydrogenase [quinone] P16083 1shj_v 0.8818 CASP7 Caspase-7 CASP7_HUMAN 1e7a_v 0.8008 ALB Serum albumin ALBU_HUMAN 2zas_v 0.7294 ESRRG Estrogen-related receptor gamma P62508 2o65_v 0.702 PIM1 Proto-oncogene serine/threonine-protein kinase Pim-1 PIM1_HUMAN 2ipw_v 0.6914 AKR1B1 Aldose reductase ALDR_HUMAN 3fzk_v 0.6896 HSPA8 Heat shock cognate 71 kDa protein P11142 1fdu_v 0.6888 HSD17B1 Estradiol 17-beta-dehydrogenase 1 P14061 1j99_v 0.6846 SULT2A1 Bile salt sulfotransferase Q06520 1b6a_v 0.6528 METAP2 Methionine aminopeptidase 2 AMPM2_HUMAN 1j78_v 0.6505 VTDB Vitamin D-binding protein VTDB_HUMAN 2zaz_v 0.5891 MAPK14 Mitogen-activated protein kinase 14 Q16539 1mkd_v 0.5789 PDE4D cAMP-specific 3,5-cyclic phosphodiesterase 4D PDE4D_HUMAN 1oiz_v 0.5723 TTPA Alpha-tocopherol transfer protein P49638 1ctr_v 0.5687 Calmodulin Calmodulin CALM_HUMAN 1ype_v 0.5659 F2 Prothrombin THRB_HUMAN 2vww_v 0.5598 EPHB4 Ephrin type-B receptor 4 EPHB4_HUMAN 2aa5_v 0.5513 NR3C2 Mineralocorticoid receptor MCR_HUMAN 2fq9_v 0.5423 CTSS Cathepsin S CATS_HUMAN 1tbf_v 0.5371 PDE5A cGMP-specific 3,5-cyclic phosphodiesterase PDE5A_HUMAN 1a28_v 0.516 PGR Progesterone receptor PRGR_HUMAN 3ddp_v 0.5137 CDK2 Cell division protein kinase 2 P24941 3ekr_v 0.5123 HSP90AA1 Heat shock protein HSP 90-alpha P07900 2uwl_v 0.5119 FA10 Coagulation factor X -

Flavone Effects on the Proteome and Transcriptome of Colonocytes in Vitro and in Vivo and Its Relevance for Cancer Prevention and Therapy
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Ernährungsphysiologie Flavone effects on the proteome and transcriptome of colonocytes in vitro and in vivo and its relevance for cancer prevention and therapy Isabel Winkelmann Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.-Prof. Dr. D. Haller Prüfer der Dissertation: 1. Univ.-Prof. Dr. H. Daniel 2. Univ.-Prof. Dr. U. Wenzel (Justus-Liebig-Universität Giessen) 3. Prof. Dr. E.C.M. Mariman (Maastricht University, Niederlande) schriftliche Beurteilung Die Dissertation wurde am 24.08.2009 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 25.11.2009 angenommen. Die Forschung ist immer auf dem Wege, nie am Ziel. (Adolf Pichler) Table of contents 1. Introduction .......................................................................................................... 1 1.1. Cancer and carcinogenesis .................................................................................. 2 1.2. Colorectal Cancer ............................................................................................... 3 1.2.1. Hereditary forms of CRC ........................................................................................ 4 1.2.2. Sporadic forms of CRC .......................................................................................... -

Thematic Review
thematic review Thematic Review Series: Proteomics An integrated omics analysis of eicosanoid biology1 Matthew W. Buczynski, Darren S. Dumlao, and Edward A. Dennis2 Department of Chemistry and Biochemistry, Department of Pharmacology, and School of Medicine, University of California, San Diego, La Jolla, CA 92093 Abstract Eicosanoids have been implicated in a vast number to address the question of how molecular biology works as of devastating inflammatory conditions, including arthritis, an integrated process (1). atherosclerosis, pain, and cancer. Currently, over a hundred Systems biology has advanced exponentially during the different eicosanoids have been identified, with many having past two decades, with transcriptomics, proteomics, and potent bioactive signaling capacity. These lipid metabolites metabolomics each playing an integral role. Each of these are synthesized de novo by at least 50 unique enzymes, many of which have been cloned and characterized. Due to the ex- platforms brings its own unique advantages and limitations tensive characterization of eicosanoid biosynthetic pathways, in facilitating the investigation of disease pathology. A this field provides a unique framework for integrating geno- transcriptomic approach can detect the upregulation and mics, proteomics, and metabolomics toward the investigation downregulation of important biosynthetic and signaling of disease pathology. To facilitate a concerted systems biol- genes; however, gene changes often donʼt directly corre- ogy approach, this review outlines the -

Product Data Sheet Purified Anti-Human CD143 (Angiotensin-Converting Enzyme)
Version: 1 Revision Date: 2012-11-30 Product Data Sheet Purified anti-human CD143 (Angiotensin-converting enzyme) Catalog # / Size: 344202 / 100 µg Clone: 5-369 Isotype: Mouse IgG1, κ Reactivity: Human Preparation: The antibody was purified by affinity chromatography. Formulation: Phosphate-buffered solution, pH 7.2, containing 0.09% sodium azide. Concentration: 0.5 mg/ml Storage: The antibody solution should be stored undiluted at 4°C. Applications: Applications: FC - Quality tested Recommended Usage: Each lot of this antibody is quality control tested by immunofluorescent staining with flow cytometric analysis. For immunofluorescent staining, the ≤ GM-CSF-stimulated human suggested use of this reagent is 0.5 µg per million cells in 100 µl volume. It monocytes (day-3) stained with the is recommended that the reagent be titrated for optimal performance for each 5-369 PE (gated in CD14+ cell application. population) Description: CD143 (ACE, kininase II, peptidyl dipeptidase 1, peptidase P, carboxycathepsin) is a 171 kD, type I, single chain transmembranal metallopeptidase, whose cofactor is zinc. Its main targets are angiotensin I and bradykinin, acting as a blood pressure regulator. CD143 is expressed in endothelial cells; varying amounts of CD143 have been reported in different epithelial cells. The activation of macrophages and histiocytes induces the expression of this molecule. Antigen References: 1. Nakamura T, et al. 2009. Int Heart J. 50:501. 2. Jayasooriya AP, et al. 2008. P. Natl. Acad. Sci. USA 105:6531. 3. Jokubaitis VJ, et al. 2008. Blood 111:4055. 4. Arndt PG, et al. 2006. J. Immunol. 177:7233. 5. Balyasnikova IV, et al. -

Epigenetics of Aging and Alzheimer's Disease
Review Epigenetics of Aging and Alzheimer’s Disease: Implications for Pharmacogenomics and Drug Response Ramón Cacabelos 1,2,* and Clara Torrellas 1,2 Received: 30 September 2015; Accepted: 8 December 2015; Published: 21 December 2015 Academic Editor: Sabrina Angelini 1 EuroEspes Biomedical Research Center, Institute of Medical Science and Genomic Medicine, 15165-Bergondo, Corunna, Spain; [email protected] 2 Chair of Genomic Medicine, Camilo José Cela University, 28692-Madrid, Spain * Correspondence: [email protected]; Tel.: +34-981-780505 Abstract: Epigenetic variability (DNA methylation/demethylation, histone modifications, microRNA regulation) is common in physiological and pathological conditions. Epigenetic alterations are present in different tissues along the aging process and in neurodegenerative disorders, such as Alzheimer’s disease (AD). Epigenetics affect life span and longevity. AD-related genes exhibit epigenetic changes, indicating that epigenetics might exert a pathogenic role in dementia. Epigenetic modifications are reversible and can potentially be targeted by pharmacological intervention. Epigenetic drugs may be useful for the treatment of major problems of health (e.g., cancer, cardiovascular disorders, brain disorders). The efficacy and safety of these and other medications depend upon the efficiency of the pharmacogenetic process in which different clusters of genes (pathogenic, mechanistic, metabolic, transporter, pleiotropic) are involved. Most of these genes are also under the influence of the epigenetic machinery. The information available on the pharmacoepigenomics of most drugs is very limited; however, growing evidence indicates that epigenetic changes are determinant in the pathogenesis of many medical conditions and in drug response and drug resistance. Consequently, pharmacoepigenetic studies should be incorporated in drug development and personalized treatments. -

Clinicopathological Examination of Dipeptidase 1 Expression in Colorectal Cancer
BIOMEDICAL REPORTS 6: 423-428, 2017 Clinicopathological examination of dipeptidase 1 expression in colorectal cancer KAZUNOSHIN TACHIBANA1, MOTONOBU SAITO1, JUN-ICHI IMAI2, EMI ITO2, YUKA YANAGISAWA3, REIKO HONMA3, KATSUHARU SAITO1, JIN ANDO1, TOMOYUKI MOMMA1, SHINJI OHKI1, TOHRU OHTAKE1, SHINYA WATANABE2, SATOSHI WAGURI4 and SEIICHI TAKENOSHITA1 1Department of Organ Regulatory Surgery; 2Medical-Industrial Translational Research Center, Fukushima Medical University School of Medicine, Fukushima 960-1295; 3Nippon Gene Co., Ltd., Tokyo 104-0054; 4Department of Anatomy and Histology, Fukushima Medical University School of Medicine, Fukushima 960-1295, Japan Received December 22, 2016; Accepted February 2, 2017 DOI: 10.3892/br.2017.870 Abstract. Dipeptidase 1 (DPEP1) is a zinc-dependent metal- Western lifestyle (1,2). Early detection and surgical resection loproteinase that is fundamental in glutathione and leukotriene is the most effective measure to improve CRC-associated metabolism. DPEP1 was initially considered as a tumor mortality (3). Despite recent advances in CRC treatment, such suppressor gene in Wilms' tumor and breast cancer. However, as chemotherapy and molecular targeted therapy, the prognosis it has been reported that DPEP1 is upregulated in colorectal remains poor in advanced CRC cases (1,2). Recurrence and cancers (CRCs) and high DPEP1 expression levels are associ- metastasis are the main obstacles for improving the prognosis ated with poorer patient survival. The role of DPEP1 genes of postoperative CRC patients (2,4). In Japan, ~15% of patients in CRC, as well as their expression, requires investigation. with stage II and 30% of patients with stage III will develop Therefore, the present study investigated DPEP1 expression recurrence within 5 years of surgical resection of CRC (5).