Cathepsin B Carboxydipeptidase Specificity Analysis Using Internally
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cystatins in Immune System
Journal of Cancer 2013, Vol. 4 45 Ivyspring International Publisher Journal of Cancer 2013; 4(1): 45-56. doi: 10.7150/jca.5044 Review Cystatins in Immune System Špela Magister1 and Janko Kos1,2 1. Jožef Stefan Institute, Department of Biotechnology, Ljubljana, Slovenia; 2. University of Ljubljana, Faculty of Pharmacy, Ljubljana, Slovenia. Corresponding author: Janko Kos, Ph. D., Department of Biotechnology, Jožef Stefan Institute, &Faculty of Pharmacy, University of Ljubljana, Slovenia; [email protected]; Phone+386 1 4769 604, Fax +3861 4258 031. © Ivyspring International Publisher. This is an open-access article distributed under the terms of the Creative Commons License (http://creativecommons.org/ licenses/by-nc-nd/3.0/). Reproduction is permitted for personal, noncommercial use, provided that the article is in whole, unmodified, and properly cited. Received: 2012.10.22; Accepted: 2012.12.01; Published: 2012.12.20 Abstract Cystatins comprise a large superfamily of related proteins with diverse biological activities. They were initially characterised as inhibitors of lysosomal cysteine proteases, however, in recent years some alternative functions for cystatins have been proposed. Cystatins pos- sessing inhibitory function are members of three families, family I (stefins), family II (cystatins) and family III (kininogens). Stefin A is often linked to neoplastic changes in epithelium while another family I cystatin, stefin B is supposed to have a specific role in neuredegenerative diseases. Cystatin C, a typical type II cystatin, is expressed in a variety of human tissues and cells. On the other hand, expression of other type II cystatins is more specific. Cystatin F is an endo/lysosome targeted protease inhibitor, selectively expressed in immune cells, suggesting its role in processes related to immune response. -

Catalytic Properties and Inhibition of Proline-Specific Dipeptidyl Peptidases II, IV and VII
UC Berkeley UC Berkeley Previously Published Works Title Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII Permalink https://escholarship.org/uc/item/4nf146zf Journal Biochemical Journal, 371 ISSN 0264-6021 Authors Leiting, B Pryor, K D Wu, J K et al. Publication Date 2003-04-01 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Biochem. J. (2003) 371, 525–532 (Printed in Great Britain) 525 Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII Barbara LEITING*1, KellyAnn D. PRYOR*, Joseph K. WU*, Frank MARSILIO*, Reshma A. PATEL*, Charles S. CRAIK†, Jonathan A. ELLMAN‡, Richard T. CUMMINGS* and Nancy A. THORNBERRY* *Department of Metabolic Disorders, Merck Research Laboratories, Mail code RY50G-236, P.O. Box 2000, Rahway, NJ 07065, U.S.A., †Department of Pharmaceutical Chemistry, University of California, 513 Parnassus Avenue, San Francisco, CA 94143-0446, U.S.A., and ‡Department of Chemistry, University of California, Berkeley, CA 94720, U.S.A. There is currently intense interest in the emerging group of strates and inhibitors for these enzymes, a complete biochemical proline-specific dipeptidases, and their roles in the regulation profile of these enzymes was obtained. The pH profiles, substrate of biological processes. Dipeptidyl peptidase IV (DPP-IV) is specificities as determined by positional scanning, Michaelis– involved in glucose metabolism by contributing to the regulation Menten constants and inhibition profiles for DPP-VII and DPP- of glucagon family peptides and has emerged as a potential target II were shown to be virtually identical, strongly supporting the for the treatment of metabolic diseases. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

JASON MARC GOLDSTEIN the Isolation, Characterization
JASON MARC GOLDSTEIN The Isolation, Characterization and Cloning of Three Novel Peptidases From Streptoccocus gordonii: Their Potential Roles in Subacute Bacterial Endocarditis (Under the Direction of JAMES TRAVIS) Streptococcus gordonii is generally considered a benign inhabitant of the oral microflora yet is a primary etiological agent in the development of subacute bacterial endocarditis (SBE), an inflammatory state that propagates thrombus formation and tissue damage on the surface of heart valves. Colonization and adherence mechanisms have been identified, yet factors necessary to sustain growth remain unidentified. Strain FSS2 produced three extracellular aminopeptidase activities during growth in neutral pH- controlled batch cultures. The first included a serine-class dipeptidyl-aminopeptidase, an x-Pro DPP (Sg-xPDPP) found as an 85 kDa monomer by SDS-PAGE while appearing as a homodimer under native conditions. Kinetic studies indicated a unique and stringent x- Pro specificity comparable to the DPPIV/CD26 and lactococcal x-Pro DPP families. Isolation of the full-length gene uncovered a 759-amino acid polypeptide with a mass of 87,115 Da and theoretical pI of 5.6. Significant homology was found with PepX gene family members from Lactobacillus ssp. and Lactococcus ssp., and putative streptococcal x-Pro DPPs. The second activity was a putative serine-class arginine aminopeptidase (Sg- RAP) with some cysteine-class characteristics. It was found as a protein monomer of 70 kDa under denaturing conditions. Nested PCR cloning enabled the isolation of a 324 bp- long DNA fragment encoding the protein’s 108 amino acid N-terminus. Culture activity profiles and N-terminal sequence analysis indicated the release of this protein from the cell surface. -

Genetic Polymorphism of the Angiotensin-Converting Enzyme (ACE) in Asthmatic Patients
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector RESPIRATORY MEDICINE (1998) 92, 1305-1310 Original Articles Genetic polymorphism of the angiotensin-converting enzyme (ACE) in asthmatic patients H. TOMITA, S. SATO, R. MATSUDA, N. OGISU, T. MORI, T. NIIMI AND S. SHIMIZU 2nd Department of Intemal Medicine, Nagoya City University Medical School, Nagoya, Japan Angiotensin-converting enzyme (ACE) inactivates bradykinin, substance P and neurokinin A, which are believed to play important roles in the pathogenesis of asthma, especially in neurogenic inflammation. It has recently been shown that an insertion (1)ldeletion (D) polymorphism in the ACE gene accounts for variation in serum ACE levels. There are thus three genotypes (insertion homozygote, II; deletion homozygote, DD; heterozygotes, DI). The serum ACE level with the DD type is reported to be about double that of the II type and intermediate in the DI case. In the present study, we examined whether asthma is linked with this ACE gene polymorphism. Seventy-one patients with asthma (27 males and 44 females) and 142 sex- and age-matched healthy controls were determined for their genotype by the polymerase chain reaction (PCR) method. Twenty-five asthmatics demonstrated the II type (352%) 37 the DI type (52.1%), and nine the DD type (12.7%). There were no significant differences in the distributions of genotypes and serum ACE levels between healthy controls and patients. No significant differences were evident in serum IgE levels, age at onset, proportion of atopic type patients and severity of asthma among the three genotypes. -

Best Presentation
Survey of Public Showed Preference for Healthcare Environ Diagnostic ment 15% Energy Healthca 13% re 47% Food & Nutrition 15% Others 11% Chagas Disease – Our Real World Problem Chagas Disease – Our Real World Problem “Chagas disease, caused by the protozoan Trypanosoma cruzi, is responsible for a greater disease burden than any other parasitic disease in the New World” Limitations in Diagnostics Immunocompromised Coinfection with HIV Infants Variable efficiency Evolution of surface antigens Differences between strains Investigating the Feasibility of Our Diagnostics Would screening all infants impact epidemiology? Would our diagnostic be a viable investment? Can our project make a real difference? Prof Yves Carlier, expert in Infectious Diseases (Université Libre de Bruxelles) Provided us with useful insights into Chagas disease throughout our project Epidemiological Model Shows a Congenital Chagas Diagnostic is Viable Total infected without diagnostic Population Total infected with diagnostic Years Epidemiological Model Shows a Congenital Chagas Diagnostic is Viable Total infected >130,000 fewer infected individuals without diagnostic $61 mil in healthcare costs saved annually Total infected with Population diagnostic 37,000 DALYs per year eliminated Years Prof Mike Bonsall, Professor of Mathematical Biology (University of Oxford) Helped us gain a better understanding of the principles of disease modelling, and equipped us with the skills to create our own epidemiological model for Chagas disease Canonical Diagnostic Circuitry INPUT CIRCUIT -

B Number Gene Name Mrna Intensity Mrna
sample) total list predicted B number Gene name assignment mRNA present mRNA intensity Gene description Protein detected - Membrane protein membrane sample detected (total list) Proteins detected - Functional category # of tryptic peptides # of tryptic peptides # of tryptic peptides detected (membrane b0002 thrA 13624 P 39 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules b0003 thrB 6781 P 9 P 3 0 homoserine kinase Metabolism of small molecules b0004 thrC 15039 P 18 P 10 0 threonine synthase Metabolism of small molecules b0008 talB 20561 P 20 P 13 0 transaldolase B Metabolism of small molecules chaperone Hsp70; DNA biosynthesis; autoregulated heat shock b0014 dnaK 13283 P 32 P 23 0 proteins Cell processes b0015 dnaJ 4492 P 13 P 4 P(m) 1 chaperone with DnaK; heat shock protein Cell processes b0029 lytB 1331 P 16 P 2 0 control of stringent response; involved in penicillin tolerance Global functions b0032 carA 9312 P 14 P 8 0 carbamoyl-phosphate synthetase, glutamine (small) subunit Metabolism of small molecules b0033 carB 7656 P 48 P 17 0 carbamoyl-phosphate synthase large subunit Metabolism of small molecules b0048 folA 1588 P 7 P 1 0 dihydrofolate reductase type I; trimethoprim resistance Metabolism of small molecules peptidyl-prolyl cis-trans isomerase (PPIase), involved in maturation of b0053 surA 3825 P 19 P 4 P(m) 1 GenProt outer membrane proteins (1st module) Cell processes b0054 imp 2737 P 42 P 5 P(m) 5 GenProt organic solvent tolerance Cell processes b0071 leuD 4770 P 10 P 9 0 isopropylmalate -

Morelloflavone and Its Semisynthetic Derivatives As Potential Novel Inhibitors of Cysteine and Serine Proteases
See discussions, stats, and author profiles for this publication at: http://www.researchgate.net/publication/276087209 Morelloflavone and its semisynthetic derivatives as potential novel inhibitors of cysteine and serine proteases ARTICLE in JOURNAL OF MEDICINAL PLANT RESEARCH · APRIL 2015 Impact Factor: 0.88 · DOI: 10.5897/JMPR2014.5641 DOWNLOADS VIEWS 2 17 8 AUTHORS, INCLUDING: Ihosvany Camps Claudio Viegas-jr Universidade Federal de Alfenas Universidade Federal de Alfenas 35 PUBLICATIONS 128 CITATIONS 6 PUBLICATIONS 22 CITATIONS SEE PROFILE SEE PROFILE Available from: Ihosvany Camps Retrieved on: 08 September 2015 Vol. 9(13), pp. 426-434, 3 April, 2015 DOI: 10.5897/JMPR2014.5641 Article Number: A42115152263 ISSN 1996-0875 Journal of Medicinal Plants Research Copyright © 2015 Author(s) retain the copyright of this article http://www.academicjournals.org/JMPR Full Length Research Paper Morelloflavone and its semisynthetic derivatives as potential novel inhibitors of cysteine and serine proteases Vanessa Silva Gontijo1, Jaqueline Pereira Januário1, Wagner Alves de Souza Júdice2, Alyne Alexandrino Antunes2, Ingridy Ribeiro Cabral1, Diego Magno Assis3, Maria Aparecida Juliano3, Ihosvany Camps4, Marcos José Marques4, Claudio Viegas Junior1 and Marcelo Henrique dos Santos1* 1Department of Exact Science, Laboratory of Phytochemistry and Medicinal Chemistry, Federal University of Alfenas, MG, Brazil. 2Interdisciplinary Center of Biochemical Investigation, Mogi das Cruzes University, Mogi das Cruzes, SP, Brazil. 3Department of Biophysics, Federal University of São Paulo, SP, Brazil. 4Department of Biological Sciences, Laboratory of Molecular Biology, Federal University of Alfenas, MG, Brazil. Received 9 October, 2014; Accepted 11 March, 2015 This article reports the three biflavonoids isolated from the fruit pericarp of Garcinia brasiliensis Mart. -

Table 6. Putative Genes Involved in the Utilization of Carbohydrates in G
Table 6. Putative genes involved in the utilization of carbohydrates in G. thermodenitrificans NG80-2 genome Carbohydrates* Enzymes Gene ID Glycerol Glycerol Kinase GT1216 Glycerol-3-phosphate dehydrogenase, aerobic GT2089 NAD(P)H-dependent glycerol-3-phosphate dehydrogenase GT2153 Enolase GT3003 2,3-bisphosphoglycerate-independentphosphoglycerate mutase GT3004 Triosephosphate isomerase GT3005 3-phosphoglycerate kinase GT3006 Glyceraldehyde-3-phosphate dehydrogenase GT3007 Phosphoglycerate mutase GT1326 Pyruvate kinase GT2663 L-Arabinose L-arabinose isomerase GT1795 L-ribulokinase GT1796 L-ribulose 5-phosphate 4-epimerase GT1797 D-Ribose Ribokinase GT3174 Transketolase GT1187 Ribose 5-phosphate epimerase GT3316 D-Xylose Xylose kinase GT1756 Xylose isomerase GT1757 D-Galactose Galactokinase GT2086 Galactose-1-phosphate uridyltransferase GT2084 UDP-glucose 4-epimerase GT2085 Carbohydrates* Enzymes Gene ID D-Fructose 1-phosphofructokinase GT1727 Fructose-1,6-bisphosphate aldolase GT1805 Fructose-1,6-bisphosphate aldolase type II GT3331 Triosephosphate isomerase GT3005 D-Mannose Mannnose-6 phospate isomelase GT3398 6-phospho-1-fructokinase GT2664 D-Mannitol Mannitol-1-phosphate dehydrogenase GT1844 N-Acetylglucosamine N-acetylglucosamine-6-phosphate deacetylase GT2205 N-acetylglucosamine-6-phosphate isomerase GT2204 D-Maltose Alpha-1,4-glucosidase GT0528, GT1643 Sucrose Sucrose phosphorylase GT3215 D-Trehalose Alpha-glucosidase GT1643 Glucose kinase GT2381 Inositol Myo-inositol catabolism protein iolC;5-dehydro-2- GT1807 deoxygluconokinase -

An Investigation on Catalysis of Acylaminoacyl Peptidases
An investigation on catalysis of acylaminoacyl peptidases PhD Thesis András László Kiss Doctorate School of Biology School leader: Prof. Anna Erdei Structural Biochemistry Programme Programme leader: Prof. László Gráf Supervisor: Prof. László Polgár Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences Budapest 2007 1. Introduction Serine peptidases contain two residues at the active site in addition to the catalytic triad (His, Asp, Ser), which form a cavity called “oxyanion hole” that accommodates the negatively charged oxyanion in the transition state of the catalysis and donate two H- bonds. Enzymes of the prolyl oligopeptidase (POP) family (acylaminoacyl peptidase, prolyl oligopeptidase, dipeptidyl peptidase IV and oligopeptidase B) are extensively studied. These enzymes are larger than classical serine peptidases and are composed of a peptidase domain with an α/β hydrolase fold and a β-propeller domain. POP and oligopeptidase B are monomeric enzymes and endopeptidases, while the exopeptidase acylaminoacyl peptidase and dipeptidyl peptidase IV are tetrameric and dimeric enzymes, respectively. Acylaminoacyl peptidase (AAP) cleaves acylated amino acids from the N-terminus of the N-acylated peptides that plays important role in many biological and disease processes. Human AAP is encoded by the DNF15S2 locus on the short arm of chromosome 3 at the region 21, which suffers deletions in small cell lung carcinomas, and renal carcinomas, resulting in deficiency in the expression of the enzyme. Acylaminoacyl peptidase is also supposed to be involved in the degradation of oxidatively damaged proteins in cells and can be associated with various diseases where damaged proteins aggregate. Its involvement in cataract formation and in the breakdown of immunogenic formylmethionyl-peptides in the digestive tract after bacterial attack was also suggested. -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -
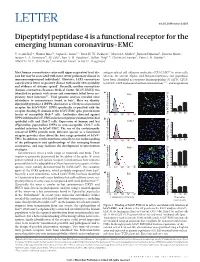
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells.