And Exopeptidases in a Processing Enzyme System: Activation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

JASON MARC GOLDSTEIN the Isolation, Characterization
JASON MARC GOLDSTEIN The Isolation, Characterization and Cloning of Three Novel Peptidases From Streptoccocus gordonii: Their Potential Roles in Subacute Bacterial Endocarditis (Under the Direction of JAMES TRAVIS) Streptococcus gordonii is generally considered a benign inhabitant of the oral microflora yet is a primary etiological agent in the development of subacute bacterial endocarditis (SBE), an inflammatory state that propagates thrombus formation and tissue damage on the surface of heart valves. Colonization and adherence mechanisms have been identified, yet factors necessary to sustain growth remain unidentified. Strain FSS2 produced three extracellular aminopeptidase activities during growth in neutral pH- controlled batch cultures. The first included a serine-class dipeptidyl-aminopeptidase, an x-Pro DPP (Sg-xPDPP) found as an 85 kDa monomer by SDS-PAGE while appearing as a homodimer under native conditions. Kinetic studies indicated a unique and stringent x- Pro specificity comparable to the DPPIV/CD26 and lactococcal x-Pro DPP families. Isolation of the full-length gene uncovered a 759-amino acid polypeptide with a mass of 87,115 Da and theoretical pI of 5.6. Significant homology was found with PepX gene family members from Lactobacillus ssp. and Lactococcus ssp., and putative streptococcal x-Pro DPPs. The second activity was a putative serine-class arginine aminopeptidase (Sg- RAP) with some cysteine-class characteristics. It was found as a protein monomer of 70 kDa under denaturing conditions. Nested PCR cloning enabled the isolation of a 324 bp- long DNA fragment encoding the protein’s 108 amino acid N-terminus. Culture activity profiles and N-terminal sequence analysis indicated the release of this protein from the cell surface. -

B Number Gene Name Mrna Intensity Mrna
sample) total list predicted B number Gene name assignment mRNA present mRNA intensity Gene description Protein detected - Membrane protein membrane sample detected (total list) Proteins detected - Functional category # of tryptic peptides # of tryptic peptides # of tryptic peptides detected (membrane b0002 thrA 13624 P 39 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules b0003 thrB 6781 P 9 P 3 0 homoserine kinase Metabolism of small molecules b0004 thrC 15039 P 18 P 10 0 threonine synthase Metabolism of small molecules b0008 talB 20561 P 20 P 13 0 transaldolase B Metabolism of small molecules chaperone Hsp70; DNA biosynthesis; autoregulated heat shock b0014 dnaK 13283 P 32 P 23 0 proteins Cell processes b0015 dnaJ 4492 P 13 P 4 P(m) 1 chaperone with DnaK; heat shock protein Cell processes b0029 lytB 1331 P 16 P 2 0 control of stringent response; involved in penicillin tolerance Global functions b0032 carA 9312 P 14 P 8 0 carbamoyl-phosphate synthetase, glutamine (small) subunit Metabolism of small molecules b0033 carB 7656 P 48 P 17 0 carbamoyl-phosphate synthase large subunit Metabolism of small molecules b0048 folA 1588 P 7 P 1 0 dihydrofolate reductase type I; trimethoprim resistance Metabolism of small molecules peptidyl-prolyl cis-trans isomerase (PPIase), involved in maturation of b0053 surA 3825 P 19 P 4 P(m) 1 GenProt outer membrane proteins (1st module) Cell processes b0054 imp 2737 P 42 P 5 P(m) 5 GenProt organic solvent tolerance Cell processes b0071 leuD 4770 P 10 P 9 0 isopropylmalate -

Table 6. Putative Genes Involved in the Utilization of Carbohydrates in G
Table 6. Putative genes involved in the utilization of carbohydrates in G. thermodenitrificans NG80-2 genome Carbohydrates* Enzymes Gene ID Glycerol Glycerol Kinase GT1216 Glycerol-3-phosphate dehydrogenase, aerobic GT2089 NAD(P)H-dependent glycerol-3-phosphate dehydrogenase GT2153 Enolase GT3003 2,3-bisphosphoglycerate-independentphosphoglycerate mutase GT3004 Triosephosphate isomerase GT3005 3-phosphoglycerate kinase GT3006 Glyceraldehyde-3-phosphate dehydrogenase GT3007 Phosphoglycerate mutase GT1326 Pyruvate kinase GT2663 L-Arabinose L-arabinose isomerase GT1795 L-ribulokinase GT1796 L-ribulose 5-phosphate 4-epimerase GT1797 D-Ribose Ribokinase GT3174 Transketolase GT1187 Ribose 5-phosphate epimerase GT3316 D-Xylose Xylose kinase GT1756 Xylose isomerase GT1757 D-Galactose Galactokinase GT2086 Galactose-1-phosphate uridyltransferase GT2084 UDP-glucose 4-epimerase GT2085 Carbohydrates* Enzymes Gene ID D-Fructose 1-phosphofructokinase GT1727 Fructose-1,6-bisphosphate aldolase GT1805 Fructose-1,6-bisphosphate aldolase type II GT3331 Triosephosphate isomerase GT3005 D-Mannose Mannnose-6 phospate isomelase GT3398 6-phospho-1-fructokinase GT2664 D-Mannitol Mannitol-1-phosphate dehydrogenase GT1844 N-Acetylglucosamine N-acetylglucosamine-6-phosphate deacetylase GT2205 N-acetylglucosamine-6-phosphate isomerase GT2204 D-Maltose Alpha-1,4-glucosidase GT0528, GT1643 Sucrose Sucrose phosphorylase GT3215 D-Trehalose Alpha-glucosidase GT1643 Glucose kinase GT2381 Inositol Myo-inositol catabolism protein iolC;5-dehydro-2- GT1807 deoxygluconokinase -

An Investigation on Catalysis of Acylaminoacyl Peptidases
An investigation on catalysis of acylaminoacyl peptidases PhD Thesis András László Kiss Doctorate School of Biology School leader: Prof. Anna Erdei Structural Biochemistry Programme Programme leader: Prof. László Gráf Supervisor: Prof. László Polgár Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences Budapest 2007 1. Introduction Serine peptidases contain two residues at the active site in addition to the catalytic triad (His, Asp, Ser), which form a cavity called “oxyanion hole” that accommodates the negatively charged oxyanion in the transition state of the catalysis and donate two H- bonds. Enzymes of the prolyl oligopeptidase (POP) family (acylaminoacyl peptidase, prolyl oligopeptidase, dipeptidyl peptidase IV and oligopeptidase B) are extensively studied. These enzymes are larger than classical serine peptidases and are composed of a peptidase domain with an α/β hydrolase fold and a β-propeller domain. POP and oligopeptidase B are monomeric enzymes and endopeptidases, while the exopeptidase acylaminoacyl peptidase and dipeptidyl peptidase IV are tetrameric and dimeric enzymes, respectively. Acylaminoacyl peptidase (AAP) cleaves acylated amino acids from the N-terminus of the N-acylated peptides that plays important role in many biological and disease processes. Human AAP is encoded by the DNF15S2 locus on the short arm of chromosome 3 at the region 21, which suffers deletions in small cell lung carcinomas, and renal carcinomas, resulting in deficiency in the expression of the enzyme. Acylaminoacyl peptidase is also supposed to be involved in the degradation of oxidatively damaged proteins in cells and can be associated with various diseases where damaged proteins aggregate. Its involvement in cataract formation and in the breakdown of immunogenic formylmethionyl-peptides in the digestive tract after bacterial attack was also suggested. -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0110747 A1 Ramseier Et Al
US 200601 10747A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0110747 A1 Ramseier et al. (43) Pub. Date: May 25, 2006 (54) PROCESS FOR IMPROVED PROTEIN (60) Provisional application No. 60/591489, filed on Jul. EXPRESSION BY STRAIN ENGINEERING 26, 2004. (75) Inventors: Thomas M. Ramseier, Poway, CA Publication Classification (US); Hongfan Jin, San Diego, CA (51) Int. Cl. (US); Charles H. Squires, Poway, CA CI2O I/68 (2006.01) (US) GOIN 33/53 (2006.01) CI2N 15/74 (2006.01) Correspondence Address: (52) U.S. Cl. ................................ 435/6: 435/7.1; 435/471 KING & SPALDING LLP 118O PEACHTREE STREET (57) ABSTRACT ATLANTA, GA 30309 (US) This invention is a process for improving the production levels of recombinant proteins or peptides or improving the (73) Assignee: Dow Global Technologies Inc., Midland, level of active recombinant proteins or peptides expressed in MI (US) host cells. The invention is a process of comparing two genetic profiles of a cell that expresses a recombinant (21) Appl. No.: 11/189,375 protein and modifying the cell to change the expression of a gene product that is upregulated in response to the recom (22) Filed: Jul. 26, 2005 binant protein expression. The process can improve protein production or can improve protein quality, for example, by Related U.S. Application Data increasing solubility of a recombinant protein. Patent Application Publication May 25, 2006 Sheet 1 of 15 US 2006/0110747 A1 Figure 1 09 010909070£020\,0 10°0 Patent Application Publication May 25, 2006 Sheet 2 of 15 US 2006/0110747 A1 Figure 2 Ester sers Custer || || || || || HH-I-H 1 H4 s a cisiers TT closers | | | | | | Ya S T RXFO 1961. -

Durham E-Theses
Durham E-Theses Midgut proteases from larval spodoptera littoralis (lepidoptera: noctutoae) Lee, Michael James How to cite: Lee, Michael James (1992) Midgut proteases from larval spodoptera littoralis (lepidoptera: noctutoae), Durham theses, Durham University. Available at Durham E-Theses Online: http://etheses.dur.ac.uk/5739/ Use policy The full-text may be used and/or reproduced, and given to third parties in any format or medium, without prior permission or charge, for personal research or study, educational, or not-for-prot purposes provided that: • a full bibliographic reference is made to the original source • a link is made to the metadata record in Durham E-Theses • the full-text is not changed in any way The full-text must not be sold in any format or medium without the formal permission of the copyright holders. Please consult the full Durham E-Theses policy for further details. Academic Support Oce, Durham University, University Oce, Old Elvet, Durham DH1 3HP e-mail: [email protected] Tel: +44 0191 334 6107 http://etheses.dur.ac.uk MIDGUT PROTEASES FROM LARVAL SPODOPTERA LITTORALIS (LEPIDOPTERA: NOCTUTOAE) By Michael James Lee B.Sc. (Dunelm) The copyright of this thesis rests with the author. No quotation from it should be pubhshed without his prior written consent and information derived from it should be acknowledged. Being a thesis submitted for the degree of Doctor of Philosophy of the University of Durham. November, 1992 Hatfield College University of Durham 6 APR 1993 DECLARATION I hereby declare that the work presented in this document is based on research carried out by me, and that no part has been previously submitted for a degree in this or any other university. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Supplementary Information
Supplementary information (a) (b) Figure S1. Resistant (a) and sensitive (b) gene scores plotted against subsystems involved in cell regulation. The small circles represent the individual hits and the large circles represent the mean of each subsystem. Each individual score signifies the mean of 12 trials – three biological and four technical. The p-value was calculated as a two-tailed t-test and significance was determined using the Benjamini-Hochberg procedure; false discovery rate was selected to be 0.1. Plots constructed using Pathway Tools, Omics Dashboard. Figure S2. Connectivity map displaying the predicted functional associations between the silver-resistant gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S3. Connectivity map displaying the predicted functional associations between the silver-sensitive gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S4. Metabolic overview of the pathways in Escherichia coli. The pathways involved in silver-resistance are coloured according to respective normalized score. Each individual score represents the mean of 12 trials – three biological and four technical. Amino acid – upward pointing triangle, carbohydrate – square, proteins – diamond, purines – vertical ellipse, cofactor – downward pointing triangle, tRNA – tee, and other – circle. -
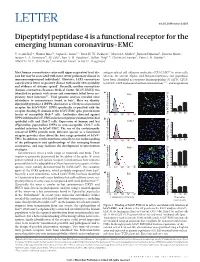
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells. -

©Ferrata Storti Foundation
Original Articles T-cell/histiocyte-rich large B-cell lymphoma shows transcriptional features suggestive of a tolerogenic host immune response Peter Van Loo,1,2,3 Thomas Tousseyn,4 Vera Vanhentenrijk,4 Daan Dierickx,5 Agnieszka Malecka,6 Isabelle Vanden Bempt,4 Gregor Verhoef,5 Jan Delabie,6 Peter Marynen,1,2 Patrick Matthys,7 and Chris De Wolf-Peeters4 1Department of Molecular and Developmental Genetics, VIB, Leuven, Belgium; 2Department of Human Genetics, K.U.Leuven, Leuven, Belgium; 3Bioinformatics Group, Department of Electrical Engineering, K.U.Leuven, Leuven, Belgium; 4Department of Pathology, University Hospitals K.U.Leuven, Leuven, Belgium; 5Department of Hematology, University Hospitals K.U.Leuven, Leuven, Belgium; 6Department of Pathology, The Norwegian Radium Hospital, University of Oslo, Oslo, Norway, and 7Department of Microbiology and Immunology, Rega Institute for Medical Research, K.U.Leuven, Leuven, Belgium Citation: Van Loo P, Tousseyn T, Vanhentenrijk V, Dierickx D, Malecka A, Vanden Bempt I, Verhoef G, Delabie J, Marynen P, Matthys P, and De Wolf-Peeters C. T-cell/histiocyte-rich large B-cell lymphoma shows transcriptional features suggestive of a tolero- genic host immune response. Haematologica. 2010;95:440-448. doi:10.3324/haematol.2009.009647 The Online Supplementary Tables S1-5 are in separate PDF files Supplementary Design and Methods One microgram of total RNA was reverse transcribed using random primers and SuperScript II (Invitrogen, Merelbeke, Validation of microarray results by real-time quantitative Belgium), as recommended by the manufacturer. Relative reverse transcriptase polymerase chain reaction quantification was subsequently performed using the compar- Ten genes measured by microarray gene expression profil- ative CT method (see User Bulletin #2: Relative Quantitation ing were validated by real-time quantitative reverse transcrip- of Gene Expression, Applied Biosystems).