Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
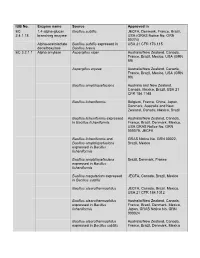
IUB No. Enzyme Name Source Approved in EC 1,4-Alpha-Glucan Bacillus Subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 Branching Enzyme USA (GRAS Notice No
IUB No. Enzyme name Source Approved in EC 1,4-alpha-glucan Bacillus subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 branching enzyme USA (GRAS Notice No. GRN 00274) Alpha-acetolactate Bacillus subtilis expressed in USA 21 CFR 173.115 decarboxylase Bacillus brevis EC 3.2.1.1 Alpha amylase Aspergillus niger Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 89) Aspergillus oryzae Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 90) Bacillus amyloliquefaciens Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Bacillus licheniformis Belgium, France, China, Japan, Denmark, Australia and New Zealand, Canada, Mexico, Brazil Bacillus licheniformis expressed Australia/New Zealand, Canada, in Bacillus licheniformis France, Brazil, Denmark, Mexico, USA GRAS Notice No. GRN 000079, JECFA Bacillus licheniformis and GRAS Notice No. GRN 00022, Bacillus amyloliquefaciens Brazil, Mexico expressed in Bacillus licheniformis Bacillus amyloliquefaciens Brazil, Denmark, France expressed in Bacillus licheniformis Bacillus megaterium expressed JECFA, Canada, Brazil, Mexico in Bacillus subtilis Bacillus stearothermophilus JECFA, Canada, Brazil, Mexico, USA 21 CFR 184.1012 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus France, Brazil, Denmark, Mexico, licheniformis Japan, GRAS Notice No. GRN 000024 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus subtilis France, Brazil, Denmark, Mexico Bacillus stearothermophilus Australia/New Zealand, Canada, (Geobacillus France, Europa, Brazil, Mexico, stearothermophilus) USA (GRN 594) Bacillus subtilis Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Rhizopus delemar Brazil Rhizopus oryzae Brazil, Mexico, USA GRAS Notice No. GRN 000090 Thermoccocales expressed in Brazil, USA (GRN126) Pseudomonas fluorecens Rhizomucor pusillus expressed Brazil, Denmark, France, Mexico, in Aspergillus niger Australia and New Zealand. -

Structure of Human Aspartyl Aminopeptidase Complexed With
Chaikuad et al. BMC Structural Biology 2012, 12:14 http://www.biomedcentral.com/1472-6807/12/14 RESEARCH ARTICLE Open Access Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family Apirat Chaikuad1, Ewa S Pilka1, Antonio De Riso2, Frank von Delft1, Kathryn L Kavanagh1, Catherine Vénien-Bryan2, Udo Oppermann1,3 and Wyatt W Yue1* Abstract Backround: Aspartyl aminopeptidase (DNPEP), with specificity towards an acidic amino acid at the N-terminus, is the only mammalian member among the poorly understood M18 peptidases. DNPEP has implicated roles in protein and peptide metabolism, as well as the renin-angiotensin system in blood pressure regulation. Despite previous enzyme and substrate characterization, structural details of DNPEP regarding ligand recognition and catalytic mechanism remain to be delineated. Results: The crystal structure of human DNPEP complexed with zinc and a substrate analogue aspartate-β- hydroxamate reveals a dodecameric machinery built by domain-swapped dimers, in agreement with electron microscopy data. A structural comparison with bacterial homologues identifies unifying catalytic features among the poorly understood M18 enzymes. The bound ligands in the active site also reveal the coordination mode of the binuclear zinc centre and a substrate specificity pocket for acidic amino acids. Conclusions: The DNPEP structure provides a molecular framework to understand its catalysis that is mediated by active site loop swapping, a mechanism likely adopted in other M18 and M42 metallopeptidases that form dodecameric complexes as a self-compartmentalization strategy. Small differences in the substrate binding pocket such as shape and positive charges, the latter conferred by a basic lysine residue, further provide the key to distinguishing substrate preference. -

(12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS Et Al
US 2016.0346364A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS et al. (43) Pub. Date: Dec. 1, 2016 (54) MEDICAMENT AND METHOD FOR (52) U.S. Cl. TREATING INNATE IMMUNE RESPONSE CPC ........... A61K 38/488 (2013.01); A61K 38/482 DISEASES (2013.01); C12Y 304/23019 (2013.01); C12Y 304/21026 (2013.01); C12Y 304/23018 (71) Applicant: DSM IPASSETS B.V., Heerlen (NL) (2013.01); A61K 9/0053 (2013.01); C12N 9/62 (2013.01); A23L 29/06 (2016.08); A2ID 8/042 (72) Inventors: Maaike Johanna BRUINS, Kaiseraugst (2013.01); A23L 5/25 (2016.08); A23V (CH); Luppo EDENS, Kaiseraugst 2002/00 (2013.01) (CH); Lenneke NAN, Kaiseraugst (CH) (57) ABSTRACT (21) Appl. No.: 15/101,630 This invention relates to a medicament or a dietary Supple (22) PCT Filed: Dec. 11, 2014 ment comprising the Aspergillus niger aspergilloglutamic peptidase that is capable of hydrolyzing plant food allergens, (86). PCT No.: PCT/EP2014/077355 and more particularly, alpha-amylase/trypsin inhibitors, thereby treating diseases due to an innate immune response S 371 (c)(1), in humans, and/or allowing to delay the onset of said (2) Date: Jun. 3, 2016 diseases. The present invention relates to the discovery that (30) Foreign Application Priority Data the Aspergillus niger aspergilloglutamic peptidase is capable of hydrolyzing alpha-amylase/trypsin inhibitors that are Dec. 11, 2013 (EP) .................................. 13196580.8 present in wheat and related cereals said inhibitors being strong inducers of innate immune response. Furthermore, Publication Classification the present invention relates to a method for hydrolyzing alpha-amylase/trypsin inhibitors comprising incubating a (51) Int. -

3 Cleavage Products of Notch 2/Site and Myelopoiesis by Dysregulating
ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch This information is current as David R. Gibb, Sheinei J. Saleem, Dae-Joong Kang, Mark of October 4, 2021. A. Subler and Daniel H. Conrad J Immunol 2011; 186:4244-4252; Prepublished online 2 March 2011; doi: 10.4049/jimmunol.1003318 http://www.jimmunol.org/content/186/7/4244 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/02/jimmunol.100331 Material 8.DC1 http://www.jimmunol.org/ References This article cites 45 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/186/7/4244.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 4, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch David R. -

ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease
International Journal of Molecular Sciences Review ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease Francesca Tosetti 1,* , Massimo Alessio 2, Alessandro Poggi 1,† and Maria Raffaella Zocchi 3,† 1 Molecular Oncology and Angiogenesis Unit, IRCCS Ospedale Policlinico S. Martino Largo R. Benzi 10, 16132 Genoa, Italy; [email protected] 2 Proteome Biochemistry, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] 3 Division of Immunology, Transplants and Infectious Diseases, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work as last author. Abstract: Enzymes, once considered static molecular machines acting in defined spatial patterns and sites of action, move to different intra- and extracellular locations, changing their function. This topological regulation revealed a close cross-talk between proteases and signaling events involving post-translational modifications, membrane tyrosine kinase receptors and G-protein coupled recep- tors, motor proteins shuttling cargos in intracellular vesicles, and small-molecule messengers. Here, we highlight recent advances in our knowledge of regulation and function of A Disintegrin And Metalloproteinase (ADAM) endopeptidases at specific subcellular sites, or in multimolecular com- plexes, with a special focus on ADAM10, and tumor necrosis factor-α convertase (TACE/ADAM17), since these two enzymes belong to the same family, share selected substrates and bioactivity. We will discuss some examples of ADAM10 activity modulated by changing partners and subcellular compartmentalization, with the underlying hypothesis that restraining protease activity by spatial Citation: Tosetti, F.; Alessio, M.; segregation is a complex and powerful regulatory tool. -

ADAMTS13 and 15 Are Not Regulated by the Full Length and N‑Terminal Domain Forms of TIMP‑1, ‑2, ‑3 and ‑4
BIOMEDICAL REPORTS 4: 73-78, 2016 ADAMTS13 and 15 are not regulated by the full length and N‑terminal domain forms of TIMP‑1, ‑2, ‑3 and ‑4 CENQI GUO, ANASTASIA TSIGKOU and MENG HUEE LEE Department of Biological Sciences, Xian Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, P.R. China Received June 29, 2015; Accepted July 15, 2015 DOI: 10.3892/br.2015.535 Abstract. A disintegrin and metalloproteinase with thom- proteolysis activities associated with arthritis, morphogenesis, bospondin motifs (ADAMTS) 13 and 15 are secreted zinc angiogenesis and even ovulation [as reviewed previously (1,2)]. proteinases involved in the turnover of von Willebrand factor Also known as the VWF-cleaving protease, ADAMTS13 and cancer suppression. In the present study, ADAMTS13 is noted for its ability in cleaving and reducing the size of the and 15 were subjected to inhibition studies with the full-length ultra-large (UL) form of the VWF. Reduction in ADAMTS13 and N-terminal domain forms of tissue inhibitor of metallo- activity from either hereditary or acquired deficiency causes proteinases (TIMPs)-1 to -4. TIMPs have no ability to inhibit accumulation of UL-VWF multimers, platelet aggregation and the ADAMTS proteinases in the full-length or N-terminal arterial thrombosis that leads to fatal thrombotic thrombocy- domain form. While ADAMTS13 is also not sensitive to the topenic purpura [as reviewed previously (1,3)]. By contrast, hydroxamate inhibitors, batimastat and ilomastat, ADAMTS15 ADAMTS15 is a potential tumor suppressor. Only a limited app can be effectively inhibited by batimastat (Ki 299 nM). In number of in-depth investigations have been carried out on the conclusion, the present results indicate that TIMPs are not the enzyme; however, expression and profiling studies have shown regulators of these two ADAMTS proteinases. -

Cell-Autonomous FLT3L Shedding Via ADAM10 Mediates Conventional Dendritic Cell Development in Mouse Spleen
Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen Kohei Fujitaa,b,1, Svetoslav Chakarovc,1, Tetsuro Kobayashid, Keiko Sakamotod, Benjamin Voisind, Kaibo Duanc, Taneaki Nakagawaa, Keisuke Horiuchie, Masayuki Amagaib, Florent Ginhouxc, and Keisuke Nagaod,2 aDepartment of Dentistry and Oral Surgery, Keio University School of Medicine, Tokyo 160-8582, Japan; bDepartment of Dermatology, Keio University School of Medicine, Tokyo 160-8582, Japan; cSingapore Immunology Network, Agency for Science, Technology and Research, Biopolis, 138648 Singapore; dDermatology Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; and eDepartment of Orthopedic Surgery, National Defense Medical College, Tokorozawa 359-8513, Japan Edited by Kenneth M. Murphy, Washington University School of Medicine, St. Louis, MO, and approved June 10, 2019 (received for review November 4, 2018) Conventional dendritic cells (cDCs) derive from bone marrow (BM) intocDC1sorcDC2stakesplaceintheBM(3),andthese precursors that undergo cascades of developmental programs to pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s terminally differentiate in peripheral tissues. Pre-cDC1s and pre- and cDC2s after migrating to nonlymphoid and lymphoid tissues. + + cDC2s commit in the BM to each differentiate into CD8α /CD103 cDCs are short-lived, and their homeostatic maintenance relies + cDC1s and CD11b cDC2s, respectively. Although both cDCs rely on on constant replenishment from the BM precursors (5). The cy- the cytokine FLT3L during development, mechanisms that ensure tokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by cDC accessibility to FLT3L have yet to be elucidated. Here, we gen- signaling through its receptor FLT3 expressed on DC precursors, erated mice that lacked a disintegrin and metalloproteinase (ADAM) is essential during the development of DCs (7, 13). -

Progress in the Field of Aspartic Proteinases in Cheese Manufacturing
Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin, Peter Dekker To cite this version: Sirma Yegin, Peter Dekker. Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering. Dairy Science & Technology, EDP sciences/Springer, 2013, 93 (6), pp.565-594. 10.1007/s13594-013-0137-2. hal-01201447 HAL Id: hal-01201447 https://hal.archives-ouvertes.fr/hal-01201447 Submitted on 17 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Dairy Sci. & Technol. (2013) 93:565–594 DOI 10.1007/s13594-013-0137-2 REVIEW PAPER Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin & Peter Dekker Received: 25 February 2013 /Revised: 16 May 2013 /Accepted: 21 May 2013 / Published online: 27 June 2013 # INRA and Springer-Verlag France 2013 Abstract Aspartic proteinases are an important class of proteinases which are widely used as milk-coagulating agents in industrial cheese production. They are available from a wide range of sources including mammals, plants, and microorganisms. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Effect of Inhibition of Matrix Metalloproteinases on Endometrial Decidualization and Implantation in Mated Rats M
Effect of inhibition of matrix metalloproteinases on endometrial decidualization and implantation in mated rats M. P. Rechtman, J. Zhang and L. A. Salamonsen Prince Henry's Institute ofMedical Research, PO Box 5152, Clayton, Victoria 3168, Australia Implantation of the embryo into the endometrium is a highly regulated event that is critical for establishment of pregnancy. Molecules involved in this process provide potential targets for post-coital contraception. The aims of this study were to determine whether matrix metalloproteinases (MMPs) are present at implantation sites in rats and whether administration of a broad-based inhibitor of MMPs could inhibit embryo implantation. Uterine extracts from non-pregnant rats and from rats on days 3\p=n-\9 of pregnancy were examined for the presence of MMPs. Doxycycline (5 or 15 mg day\m=-\1) was administered by gavage to rats from the day of mating (day 0) to day 7 of pregnancy and the uterus was examined for implantation sites. A number of MMPs were present in all uterine samples. MMP-2 reached a peak on day 3, whereas the highest expression of MMP-7 occurred on day 7. MMP-13 and MMP-3 were present in smaller amounts. MMP-9 was detectable only on day 9. Treatment of rats with doxycycline had no effect on the number of implantation sites or on the total uterine mass. However, in treated rats, the process of decidualization was impaired and both the width and length of the decidual zone was reduced, resulting in a decrease in total decidual area from 1.20 \m=+-\0.07 to 0.91 \m=+-\0.07 mm2 (mean \m=+-\sem, controls versus doxycycline treated, P < 0.02). -

B Number Gene Name Mrna Intensity Mrna
sample) total list predicted B number Gene name assignment mRNA present mRNA intensity Gene description Protein detected - Membrane protein membrane sample detected (total list) Proteins detected - Functional category # of tryptic peptides # of tryptic peptides # of tryptic peptides detected (membrane b0002 thrA 13624 P 39 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules b0003 thrB 6781 P 9 P 3 0 homoserine kinase Metabolism of small molecules b0004 thrC 15039 P 18 P 10 0 threonine synthase Metabolism of small molecules b0008 talB 20561 P 20 P 13 0 transaldolase B Metabolism of small molecules chaperone Hsp70; DNA biosynthesis; autoregulated heat shock b0014 dnaK 13283 P 32 P 23 0 proteins Cell processes b0015 dnaJ 4492 P 13 P 4 P(m) 1 chaperone with DnaK; heat shock protein Cell processes b0029 lytB 1331 P 16 P 2 0 control of stringent response; involved in penicillin tolerance Global functions b0032 carA 9312 P 14 P 8 0 carbamoyl-phosphate synthetase, glutamine (small) subunit Metabolism of small molecules b0033 carB 7656 P 48 P 17 0 carbamoyl-phosphate synthase large subunit Metabolism of small molecules b0048 folA 1588 P 7 P 1 0 dihydrofolate reductase type I; trimethoprim resistance Metabolism of small molecules peptidyl-prolyl cis-trans isomerase (PPIase), involved in maturation of b0053 surA 3825 P 19 P 4 P(m) 1 GenProt outer membrane proteins (1st module) Cell processes b0054 imp 2737 P 42 P 5 P(m) 5 GenProt organic solvent tolerance Cell processes b0071 leuD 4770 P 10 P 9 0 isopropylmalate -

Methionine Aminopeptidase Emerging Role in Angiogenesis
Chapter 2 Methionine Aminopeptidase Emerging role in angiogenesis Joseph A. Vetro1, Benjamin Dummitt2, and Yie-Hwa Chang2 1Department of Pharmaceutical Chemistry, University of Kansas, 2095 Constant Ave., Lawrence, KS 66047, USA. 2Edward A. Doisy Department of Biochemistry and Molecular Biology, St. Louis University Health Sciences Center, 1402 S. Grand Blvd., St. Louis, MO 63104, USA. Abstract: Angiogenesis, the formation of new blood vessels from existing vasculature, is a key factor in a number of vascular-related pathologies such as the metastasis and growth of solid tumors. Thus, the inhibition of angiogenesis has great potential as a therapeutic modality in the treatment of cancer and other vascular-related diseases. Recent evidence suggests that the inhibition of mammalian methionine aminopeptidase type 2 (MetAP2) catalytic activity in vascular endothelial cells plays an essential role in the pharmacological activity of the most potent small molecule angiogenesis inhibitors discovered to date, the fumagillin class. Methionine aminopeptidase (MetAP, EC 3.4.11.18) catalyzes the non-processive, co-translational hydrolysis of initiator N-terminal methionine when the second residue of the nascent polypeptide is small and uncharged. Initiator Met removal is a ubiquitous and essential modification. Indirect evidence suggests that removal of initiator Met by MetAP is important for the normal function of many proteins involved in DNA repair, signal transduction, cell transformation, secretory vesicle trafficking, and viral capsid assembly and infection. Currently, much effort is focused on understanding the essential nature of methionine aminopeptidase activity and elucidating the role of methionine aminopeptidase type 2 catalytic activity in angiogenesis. In this chapter, we give an overview of the MetAP proteins, outline the importance of initiator Met hydrolysis, and discuss the possible mechanism(s) through which MetAP2 inhibition by the fumagillin class of angiogenesis inhibitors leads to cytostatic growth arrest in vascular endothelial cells.