Tagzyme™ Handbook
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

JASON MARC GOLDSTEIN the Isolation, Characterization
JASON MARC GOLDSTEIN The Isolation, Characterization and Cloning of Three Novel Peptidases From Streptoccocus gordonii: Their Potential Roles in Subacute Bacterial Endocarditis (Under the Direction of JAMES TRAVIS) Streptococcus gordonii is generally considered a benign inhabitant of the oral microflora yet is a primary etiological agent in the development of subacute bacterial endocarditis (SBE), an inflammatory state that propagates thrombus formation and tissue damage on the surface of heart valves. Colonization and adherence mechanisms have been identified, yet factors necessary to sustain growth remain unidentified. Strain FSS2 produced three extracellular aminopeptidase activities during growth in neutral pH- controlled batch cultures. The first included a serine-class dipeptidyl-aminopeptidase, an x-Pro DPP (Sg-xPDPP) found as an 85 kDa monomer by SDS-PAGE while appearing as a homodimer under native conditions. Kinetic studies indicated a unique and stringent x- Pro specificity comparable to the DPPIV/CD26 and lactococcal x-Pro DPP families. Isolation of the full-length gene uncovered a 759-amino acid polypeptide with a mass of 87,115 Da and theoretical pI of 5.6. Significant homology was found with PepX gene family members from Lactobacillus ssp. and Lactococcus ssp., and putative streptococcal x-Pro DPPs. The second activity was a putative serine-class arginine aminopeptidase (Sg- RAP) with some cysteine-class characteristics. It was found as a protein monomer of 70 kDa under denaturing conditions. Nested PCR cloning enabled the isolation of a 324 bp- long DNA fragment encoding the protein’s 108 amino acid N-terminus. Culture activity profiles and N-terminal sequence analysis indicated the release of this protein from the cell surface. -

Methionine Aminopeptidase Emerging Role in Angiogenesis
Chapter 2 Methionine Aminopeptidase Emerging role in angiogenesis Joseph A. Vetro1, Benjamin Dummitt2, and Yie-Hwa Chang2 1Department of Pharmaceutical Chemistry, University of Kansas, 2095 Constant Ave., Lawrence, KS 66047, USA. 2Edward A. Doisy Department of Biochemistry and Molecular Biology, St. Louis University Health Sciences Center, 1402 S. Grand Blvd., St. Louis, MO 63104, USA. Abstract: Angiogenesis, the formation of new blood vessels from existing vasculature, is a key factor in a number of vascular-related pathologies such as the metastasis and growth of solid tumors. Thus, the inhibition of angiogenesis has great potential as a therapeutic modality in the treatment of cancer and other vascular-related diseases. Recent evidence suggests that the inhibition of mammalian methionine aminopeptidase type 2 (MetAP2) catalytic activity in vascular endothelial cells plays an essential role in the pharmacological activity of the most potent small molecule angiogenesis inhibitors discovered to date, the fumagillin class. Methionine aminopeptidase (MetAP, EC 3.4.11.18) catalyzes the non-processive, co-translational hydrolysis of initiator N-terminal methionine when the second residue of the nascent polypeptide is small and uncharged. Initiator Met removal is a ubiquitous and essential modification. Indirect evidence suggests that removal of initiator Met by MetAP is important for the normal function of many proteins involved in DNA repair, signal transduction, cell transformation, secretory vesicle trafficking, and viral capsid assembly and infection. Currently, much effort is focused on understanding the essential nature of methionine aminopeptidase activity and elucidating the role of methionine aminopeptidase type 2 catalytic activity in angiogenesis. In this chapter, we give an overview of the MetAP proteins, outline the importance of initiator Met hydrolysis, and discuss the possible mechanism(s) through which MetAP2 inhibition by the fumagillin class of angiogenesis inhibitors leads to cytostatic growth arrest in vascular endothelial cells. -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -

Effect of Secretory Particles in Bovine Seminal Vesicle Secretion on Sperm Motility and Acrosome Reaction Y
Effect of secretory particles in bovine seminal vesicle secretion on sperm motility and acrosome reaction Y. Agrawal and T. Vanha-Perttula Department of Anatomy, University of Kuopio, P.O. Box 6, 70211 Kuopio, Finland Summary. Particles found in bovine seminal vesicle secretion were enriched by centrifu- gation. They varied in size and morphology and contained Mg2+,Ca2+-activated ATPase, aminopeptidase A, alanyl aminopeptidase, \g=g\-glutamyltranspeptidase and dipeptidyl peptidase IV activities. Hyperactivation of sperm motility and the acrosome reaction were induced by these particles in epididymal spermatozoa suspended in a modified Ringer medium. The hyperactivation, analysed with a microscopic slide test, started within minutes of exposure to membrane particles and continued for 3\p=n-\4h, during which time spermatozoa underwent the acrosome reaction. Acrosome staining, phase-contrast microscopy and transmission electron microscopy revealed that the acrosome reaction started within 60 min at 37\s=deg\Cand affected up to 80% of sperm- atozoa in 4 h. These membrane particles differed from those reported previously in other species in enzyme composition, function and organ of origin. Introduction Seminal fluid comprises the secretions of the testis, epididymis and sex accessory glands. The com¬ ponents of the seminal fluid can influence the motility and metabolism of spermatozoa (Inskeep et al, 1985) with beneficial as well as deleterious effects being reported (Eliasson & Lindholmer, 1975; Dott et al, 1979; Mann & Lutwak-Mann, 1981; Baas et al, 1983). Metz et al (1968) and Davis (1973) reported the presence of membrane particles in the seminal plasma of rabbit, bull and man. Ronquist et al (1978a, b) described similar particles in the seminal fluid and prostatic secretion of man and called them prostasomes on the basis of their obvious origin from the prostate (Brody et al, 1983). -

Durham E-Theses
Durham E-Theses Midgut proteases from larval spodoptera littoralis (lepidoptera: noctutoae) Lee, Michael James How to cite: Lee, Michael James (1992) Midgut proteases from larval spodoptera littoralis (lepidoptera: noctutoae), Durham theses, Durham University. Available at Durham E-Theses Online: http://etheses.dur.ac.uk/5739/ Use policy The full-text may be used and/or reproduced, and given to third parties in any format or medium, without prior permission or charge, for personal research or study, educational, or not-for-prot purposes provided that: • a full bibliographic reference is made to the original source • a link is made to the metadata record in Durham E-Theses • the full-text is not changed in any way The full-text must not be sold in any format or medium without the formal permission of the copyright holders. Please consult the full Durham E-Theses policy for further details. Academic Support Oce, Durham University, University Oce, Old Elvet, Durham DH1 3HP e-mail: [email protected] Tel: +44 0191 334 6107 http://etheses.dur.ac.uk MIDGUT PROTEASES FROM LARVAL SPODOPTERA LITTORALIS (LEPIDOPTERA: NOCTUTOAE) By Michael James Lee B.Sc. (Dunelm) The copyright of this thesis rests with the author. No quotation from it should be pubhshed without his prior written consent and information derived from it should be acknowledged. Being a thesis submitted for the degree of Doctor of Philosophy of the University of Durham. November, 1992 Hatfield College University of Durham 6 APR 1993 DECLARATION I hereby declare that the work presented in this document is based on research carried out by me, and that no part has been previously submitted for a degree in this or any other university. -
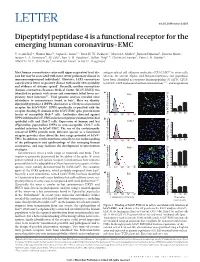
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells. -

And Exopeptidases in a Processing Enzyme System: Activation
Proc. Nail. Acad. Sci. USA Vol. 85, pp. 5468-5472, August 1988 Biochemistry Relationship between endo- and exopeptidases in a processing enzyme system: Activation of an endoprotease by the aminopeptidase B-like activity in somatostatin-28 convertase (brain cortex/basic amino acid pairs/peptide substrates/protease inhibitors/prohormone maturation) SOPHIE GOMEZ, PABLO GLUSCHANKOF, AGNES LEPAGE, AND PAUL COHEN Groupe de Neurobiochimie Cellulaire et Moldculaire, Universitd Pierre et Marie Curie, Unitd Associde 554 au Centre National de la Recherche Scientifique, 96 boulevard Raspail, 75006 Paris, France Communicated by I. Robert Lehman, April 8, 1988 (receivedfor review December 15, 1987) ABSTRACT The somatostatin-28 convertase activity in- somatostatin-14 and the amino-terminal dodecapeptide so- volved in vitro in the processing of somatostatin-28 into the matostatin-28-(1-12) (16). neuropeptides somatostatin-28-(1-12) and somatostatin-14 is We have described an endoprotease that cleaves the composed of an endoprotease and a basic aminopeptidase. We peptide bond on the amino side ofthe Arg-Lys doublet in the report herein on the purification to apparent homogeneity of somatostatin-28 sequence (17, 18), releasing the somato- these two constituents and on their functional interrelationship. statin-28-(1-12) fragment and [Arg-2,Lys-1]somatostatin-14. In particular we observed that after various physicochemical The released [Arg-2,Lys-']somatostatin-14 is further proc- treatments, the 90-kDa endoprotease activity was recovered essed by an aminopeptidase B-like activity (18, 19) that is also both at this molecular mass and as a 45-kDa entity. Moreover, present in the preparation. We report herein the purification the production of [Arg2,LysJllsomatostatin-14 from somato- to apparent homogeneity ofthese two activities. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -

Proteolytic Cleavage—Mechanisms, Function
Review Cite This: Chem. Rev. 2018, 118, 1137−1168 pubs.acs.org/CR Proteolytic CleavageMechanisms, Function, and “Omic” Approaches for a Near-Ubiquitous Posttranslational Modification Theo Klein,†,⊥ Ulrich Eckhard,†,§ Antoine Dufour,†,¶ Nestor Solis,† and Christopher M. Overall*,†,‡ † ‡ Life Sciences Institute, Department of Oral Biological and Medical Sciences, and Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada ABSTRACT: Proteases enzymatically hydrolyze peptide bonds in substrate proteins, resulting in a widespread, irreversible posttranslational modification of the protein’s structure and biological function. Often regarded as a mere degradative mechanism in destruction of proteins or turnover in maintaining physiological homeostasis, recent research in the field of degradomics has led to the recognition of two main yet unexpected concepts. First, that targeted, limited proteolytic cleavage events by a wide repertoire of proteases are pivotal regulators of most, if not all, physiological and pathological processes. Second, an unexpected in vivo abundance of stable cleaved proteins revealed pervasive, functionally relevant protein processing in normal and diseased tissuefrom 40 to 70% of proteins also occur in vivo as distinct stable proteoforms with undocumented N- or C- termini, meaning these proteoforms are stable functional cleavage products, most with unknown functional implications. In this Review, we discuss the structural biology aspects and mechanisms -
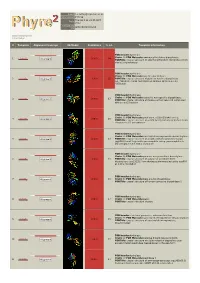
Phyre 2 Results for P15288
Email [email protected] Description P15288 Thu Jan 5 11:34:45 GMT Date 2012 Unique Job d2087d8d303c52c9 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:hydrolase Chain: B: PDB Molecule:aminoacyl-histidine dipeptidase; 1 c3mruB_ 100.0 64 Alignment PDBTitle: crystal structure of aminoacylhistidine dipeptidase from vibrio2 alginolyticus PDB header:hydrolase Chain: B: PDB Molecule:xaa-his dipeptidase; 2 c2qyvB_ Alignment 100.0 55 PDBTitle: crystal structure of putative xaa-his dipeptidase (yp_718209.1) from2 haemophilus somnus 129pt at 2.11 a resolution PDB header:hydrolase Chain: A: PDB Molecule:cytosolic non-specific dipeptidase; 3 c2zogA_ 100.0 17 Alignment PDBTitle: crystal structure of mouse carnosinase cn2 complexed with zn and2 bestatin PDB header:hydrolase Chain: B: PDB Molecule:peptidase, m20/m25/m40 family; 4 c2pokB_ 100.0 16 Alignment PDBTitle: crystal structure of a m20 family metallo peptidase from streptococcus2 pneumoniae PDB header:hydrolase Chain: A: PDB Molecule:succinyl-diaminopimelate desuccinylase; 5 c3pfeA_ Alignment 100.0 13 PDBTitle: crystal structure of a m20a metallo peptidase (dape, lpg0809) from2 legionella pneumophila subsp. pneumophila str. philadelphia 1 at 1.503 a resolution PDB header:hydrolase Chain: B: PDB Molecule:putative acetylornithine deacetylase; 6 c3pfoB_ Alignment 100.0 16 PDBTitle: crystal structure of a putative acetylornithine deacetylase (rpa2325)2 from rhodopseudomonas palustris cga009 at 1.90 a resolution PDB -

Contribution of Endo- and Exopeptidases to Formation of Non-Protein N During Ensiling of Alfalfa
Contribution of endo- and exopeptidases to formation of non-protein N during ensiling of alfalfa Dr. Xusheng Guo,1 W. Cheng,1 L. Tao,2 Yu Zhu,2 F.Y. Yang 2 & H. Zhou2 1.School of Life Science, Lanzhou University 1. International Centre for Tibetan Plateau Ecosystem Management (ICTPEM), Lanzhou University 2. Institute of Grassland Science, China Agricultural University Hämeenlinna, Finland 2-4 July 2012 Introduction Alfalfa (Medicago Sativa L.) is well known for its high nutritive value However, after ensiling: N use efficiency Extensive ppyroteolysis Reduce True protein NPN(Peptide, FAA, NH3-N etc.) Silage DM intake (44-87% of Total N; Muck, 1987) Silage Fermentation Proteolysis in ensiled forage mainly results from plant proteases (Ohshima and McDonald, 1978; McKersie, 1981; Heron et al., 1988). Proteases (peptidases) are divided into 2 classes (NC-IUBMB, 1992): Exopeptidase Endopeptidase Objectives Proteases (peptidases, E.C.3.4) Endopeptidases Exopeptidases ?! Ser ine pep tidase (E .C .3 .4 .21) Aminopeptidase (EC 3.4.11) Carboxypeptidase (EC 3.4.16) Cysteine peptidase (E.C.3.4.22) Dipeptidase (EC 3.4.13) Aspartic Peptidase (E.C.3.4.23) Dipeptidyl-peptidase (EC 3.4.14) Metallopeptidase (E.C.3.4.24) Tripeptidyl-peptidase (EC 3.4.14) Peptidyl-dipeptidase (EC 3.4.15) Aims of our research were: 1. To clarify the classes of exo- and endopeptidases that are involved in proteolysis within ensiled alfalfa. 2. To determine the contribution of these peptidases to the formation of different NPN compounds (peptide-N, FAA-N, and NH3-N) during