Msc Biochemistry Autonomy 2015-17
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
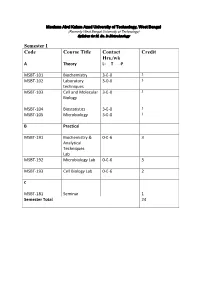
Syllabus for M
Maulana Abul Kalam Azad University of Technology, West Bengal (Formerly West Bengal University of Technology) Syllabus for M. Sc. In Biotechnology Semester I Code Course Title Contact Credit Hrs./wk A Theory L- T -P MSBT-101 Biochemistry 3-0-0 3 MSBT-102 Laboratory 3-0-0 3 techniques MSBT-103 Cell and Molecular 3-0-0 3 Biology MSBT-104 Biostatistics 3-0-0 3 MSBT-105 Microbiology 3-0-0 3 B Practical MSBT-191 Biochemistry & 0-0-6 3 Analytical Techniques Lab MSBT-192 Microbiology Lab 0-0-6 3 MSBT-193 Cell Biology Lab 0-0-6 2 C MSBT-181 Seminar 1 Semester Total 24 Maulana Abul Kalam Azad University of Technology, West Bengal (Formerly West Bengal University of Technology) Syllabus for M. Sc. In Biotechnology MSBT101: Biochemistry credits 3 Unit 1: Basic chemistry for biologists Formation of chemical bonds, molecular orbital (MO) theory and linear combination of atomic orbitals (LCAO),basics of mass spectrometry, molecules, Avogadro number, molarity, chemical reactions, reaction stoichiometry, rates of reaction, rate constants, order of reactions,kinetic versus thermodynamic controls of a reaction, reaction equilibrium (equilibrium constant); light and matter interactions (optical spectroscopy, fluorescence, bioluminescence, paramagnetism and diamagnetism, photoelectron spectroscopy; chemical bonds (ionic, covalent, Van derWalls forces); electronegativity, polarity; VSEPR theory and molecular geometry, dipole moment, orbital hybridizations; acids, bases and pH - Arrhenious theory, pH, ionic product of water, weak acids and bases, conjugate acid-base pairs, buffers and buffering action etc; chemical thermodynamics - internal energy, heat and temperature, enthalpy (bond enthalpy and reaction enthalpy), entropy, Gibbs free energy of ATP driven reactions, spontaneity versus driven reactions in biology;bond rotations and molecular conformations - Newman projections, conformational analysis of alkanes, alkenes and alkynes; functional groups, optically asymmetric carbon centers, amino acids, proteins, rotational freedoms in polypeptide backbone (Ramachandran plot). -

Technical Methods
J Clin Pathol 1987;40:581-588 J Clin Pathol: first published as 10.1136/jcp.40.5.581 on 1 May 1987. Downloaded from 56°C for 30 minutes. Technical methods Complement fixation tests were performed accord- ing to established methods,10 1 except that microtitre plates were used instead of World Health Organisation trays. For maximum sensitivity an ini- Cytomegalovirus (CMV) tial serum dilution of 1/4 was used. The antigen prep- antibody screening in blood aration used was a CMV complement fixation test antigen supplied by either Flow Laboratories Ltd, donors: modification of new latex Irvine, Scotland, or the Central Public Health Labo- ratory, Colindale, England. Guinea pig complements agglutination test compared with were supplied by Wellcome Diagnostics, Dartford, two standard methods England, or Don Whitly Scientific Ltd, Shipley, England. Complement fixation tests were performed A PUCKETT J E DAVIS From the Regional Blood using the following CMV antigen and complement Transfusion Centre, John Radeliffe Hospital, combinations: (1) PHLS CMV antigen + Wellcome Headington, Oxford, England Diagnostics complement, (2) PHLS CMV antigen + Don Whitly complement, and (3) Flow Laboratories CMV antigen + Wellcome Diagnostics complement. Infection with cytomegalovirus (CMV) is common, Immunofluorescence tests were performed and between 50 and 100% of adults may show evi- according to a standard method12 13 using substrate dence of infection.1 The transmission of the virus by slides of CMV infected (Westwood strain) fibroblasts blood transfusion2 and, therefore, the need to screen the Oxford Public Health Laboratory. donations intended for at risk groups such as provided by immunocompromised patients34 and neonates5 -7 iS CMV Scan passive latex agglutination kits were now well established. -

Serological Methods in the Identification and Characterization of Viruses
CHAPTER 4 Serological Methods in the Identification and Characterization of Viruses M. H. V. Van Regenmortel Laboratoire de Virologie Institut de Biologie Mo!eculaire et Cellulaire 67000 Strasbourg, France 1. INTRODUCTION The purpose of this chapter is to present an integrated view of the various serological techniques that have been used in virology. The accent will be placed on the principles that govern each type of test and on the general applicability of the different serological techniques in all fields of virus research. In recent years, advances in serological tech niques have sometimes been applied in only one area of virology, although they could have been equally useful to workers studying other groups of viruses. No doubt this stems from the host-oriented approach that has guided the compartmentation of virology into separate fields of specialization. When it comes to serological properties, however, the similarities between animal, insect, bacterial, and plant viruses are paramount. The same immunochemical principles govern the in vitro serological reactions of all viral antigens, and much of general interest can be learned from the findings obtained with each particular group of viruses. An attempt will be made here to emphasize the general validity of specific experimental procedures. A number of recent reviews restricted to the serology of particular groups of viruses are available 183 H. Fraenkel-Conrat et al. (eds.), Comprehensive Virology © Plenum Press, New York 1981 184 Chapter 4 (Cowan, 1973; Schmidt and Lennette, 1973; Ball, 1974; Kurstak and Morisset, 1974; Burns and Allison, 1975; Mazzone and Tignor, 1976; Mayr et al., 1977; Tyrrell, 1978; Van Regenmortel, 1978; Cooper, 1979). -

I M M U N O L O G Y Core Notes
II MM MM UU NN OO LL OO GG YY CCOORREE NNOOTTEESS MEDICAL IMMUNOLOGY 544 FALL 2011 Dr. George A. Gutman SCHOOL OF MEDICINE UNIVERSITY OF CALIFORNIA, IRVINE (Copyright) 2011 Regents of the University of California TABLE OF CONTENTS CHAPTER 1 INTRODUCTION...................................................................................... 3 CHAPTER 2 ANTIGEN/ANTIBODY INTERACTIONS ..............................................9 CHAPTER 3 ANTIBODY STRUCTURE I..................................................................17 CHAPTER 4 ANTIBODY STRUCTURE II.................................................................23 CHAPTER 5 COMPLEMENT...................................................................................... 33 CHAPTER 6 ANTIBODY GENETICS, ISOTYPES, ALLOTYPES, IDIOTYPES.....45 CHAPTER 7 CELLULAR BASIS OF ANTIBODY DIVERSITY: CLONAL SELECTION..................................................................53 CHAPTER 8 GENETIC BASIS OF ANTIBODY DIVERSITY...................................61 CHAPTER 9 IMMUNOGLOBULIN BIOSYNTHESIS ...............................................69 CHAPTER 10 BLOOD GROUPS: ABO AND Rh .........................................................77 CHAPTER 11 CELL-MEDIATED IMMUNITY AND MHC ........................................83 CHAPTER 12 CELL INTERACTIONS IN CELL MEDIATED IMMUNITY ..............91 CHAPTER 13 T-CELL/B-CELL COOPERATION IN HUMORAL IMMUNITY......105 CHAPTER 14 CELL SURFACE MARKERS OF T-CELLS, B-CELLS AND MACROPHAGES...............................................................111 -

Comparison Between Haemagglutination Inhibition and Complement Fixation Tests in Detecting Antibodies Responses Following Influenza Viral Infection
Egypt. Acad. J. Biolog. Sci., 4(1): 35-38 (2012) G. Microbiology Email:[email protected] ISSN: 2090-0872 Received: 20/7/2012 www.eajbs.eg.net Comparison between Haemagglutination Inhibition and Complement Fixation Tests in Detecting Antibodies Responses Following Influenza Viral Infection Muthana, A.K. Al-zobaei Medical Microbiology, Microbiol. Dept. Virology, College of Medicine, Univ. Anbar. ABSTRACT Objectives: To evaluate sensitivity and specificity of HI and CF tests for detecting antibody responses following naturally occurring influenza A and B infections. Methods: One hundred and twenty blood samples were drawn from one hundred patients with flue-like illness and twenty apparently healthy control subjects and tested at Central Health Laboratory in Baghdad/ Iraq using HI and CF tests for detecting three currently circulating influenza A and B strains during the period from the 1st of January 2008 to the 1st of June 2009. Results: 78-80% of the patients with flu-like illness responded to a t least one of three influenza virus antigens as measured by HI, while 34-35% of those patients showed a response by CF test. Conclusions: HI test has a higher sensitivity and lower specificity than CF test for detecting antibody responses from patients with influenza A and B viral infections. Keywords : Influenza viruses, HI, CF, Influenza serology INTRODUCTION SD. (1986), whereas CF test based on Several studies have clearly fixation of complement by antigen- demonstrated that HI is a more sensitive antibody complexes. A known amount of method than CF test for detecting complement is included in the test and antibody responses to naturally occurring residual complement is detected by the influenza virus A and B infections, addition of sheep RBCs sensitized with Julkunen, I. -
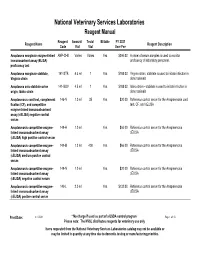
NVSL Reagent Manual
National Veterinary Services Laboratories Reagent Manual Reagent Amount/ Tests/ Billable FY 2021 Reagent Name Reagent Description Code Vial Vial User Fee Anaplasma marginale enzyme-linked ANP-CHK Varies VariesYes $394.00 A panel of serum samples is used to monitor immunosorbent assay (ELISA) proficiency of laboratory personnel. proficiency test Anaplasma marginale stabilate, 141-STB 4.5 ml 1Yes $188.00 Virginia strain, stabilate is used to initiate infection in Virginia strain donor animals Anaplasma ovis stabilate ovine 141-SOV 4.5 ml 1Yes $188.00 Idaho strain - stabilate is used to initate infection in origin, Idaho strain donor animals Anaplasmosis card test, complement 146-N 1.0 ml 35Yes $20.00 Reference control serum for the Anaplasmosis card fixation (CF), and competitive test, CF, and cELISA enzyme-linked immunoabsorbent assay (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-H 1.0 mlYes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) high positive control serum Anaplasmosis competitive enzyme- 149-M 1.0 ml 400Yes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) medium positve control serum Anaplasmosis competitive enzyme- 149-N 1.0 mlYes $20.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-L 2.0 mlYes $132.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) positve control serum Print Date: 6/11/2021 * No charge if used as part of a USDA control program Page 1 of 58 Please note: The NVSL distributes reagents for veterinary use only Items requested from the National Veterinary Services Laboratories catalog may not be available or may be limited in quantity at any time due to domestic testing or manufacturing priorities. -

Clinical Reactivity and Immunogenicity of Hemagglutinin Influenza Vaccine
Archiv fiir die gesamte Virusforschung 36, 43--52 (1972) by Springer-Verlag 1972 Clinical Reactivity and Immunogenicity of Hemagglutinin Influenza Vaccine I. Clinical Reactions, Hemagghtination-Inhibiting and Strain and Type-Specific Complement-Fixing Antibody Responses in Subjects Aged 3--6, 16--17 and 27--50 Years By HANA ZJ~VADOVX, VLADIM~I/r VONKA, EnvI~ ADAM, EvA DOMOR•ZKOV•, and FRED M. DAVENPORT1 Department of Experimental Virology and Clinical and Epidemiology Department, Institute of Sera and Vaccines, Prague, Czechoslovakia Received April 5, 1971 Summary Subjects aged 3--6, 16--17 and 27--50 years were vaccinated with one dose of hemagglutinin influenza virus vaccine. Clinical reactions, hemagglutination- inhibiting (HI) and strain- and type-specific complement-fixing (CF-V and CF-S) antibodies were determined in sera taken before and four weeks after vaccine administration. The results indicated that the reactogenicity of the vaccine was very low. The HI antibody response differed With the age of the vaceinees, apparently being conditioned by prior exposure to the various influenza virus subtypes. The results of CF tests using strain-specific V antigens corresponded in general with HI tests, with two marked exceptions. In the youngest group nearly half of the subjects developed CF antibody to V-Swine, while all of them remained without antibody detectable in the HI test. However, when V antigen was used instead of intact virus as hemagglutinin, the post-vaccination sera of these subjects also reacted positively in the HI test. Secondly, a number of pre- vaccination sera from persons aged 27--50 years possessed CF antibody to A/PR 8 in the: absence of homologous HI antibody. -

Immunology and Serology
LECTURE NOTES For Medical Laboratory Technology Students Immunology and Serology Selamawit Debebe Alemaya University In collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education 2004 Funded under USAID Cooperative Agreement No. 663-A-00-00-0358-00. Produced in collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education. Important Guidelines for Printing and Photocopying Limited permission is granted free of charge to print or photocopy all pages of this publication for educational, not-for-profit use by health care workers, students or faculty. All copies must retain all author credits and copyright notices included in the original document. Under no circumstances is it permissible to sell or distribute on a commercial basis, or to claim authorship of, copies of material reproduced from this publication. ©2004 by Selamawit Debebe All rights reserved. Except as expressly provided above, no part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system, without written permission of the author or authors. This material is intended for educational use only by practicing health care workers or students and faculty in a health care field. Immunology and Serology Preface Immunology and serology is an advanced science dealing with how the human immune system organized, function and the different types of serological techniques. It is a very vast subject covering a wide area of technology. -

Glossary 298 Appendix A
Appendix A Glossary 298 Appendix A A Angstrom units (lO-IOm)_ and/or of short duration; or a disease that has a rapid onset. aa amino acid. severe symptoms and brief duration. See also chronic. Ab antibody. ADCC antibody-dependent cell-mediated cytotoxicity. ABC antigen binding protein. addiction See dependence. ABO A system of human blood groups. additive response Where administration of two drugs ABPI Association of the British Pharmaceutical Industry. produces a response that is the simple sum of their individ absorption In pharmacology. the uptake of a drug from ual responses, i.e. there is not synergism or antagonism. its site of administration. adenyl cyclase See adenylyl cyclase. abundance The term used to describe the average adenyl ate cyclase See adenylyl cyclase. number of molecules of a particular mRNA per cell. adenylyl cyclase The enzyme that produces the second abuse liability of drugs The propensity of a drug to messenger cyclic-AMP from ATP. lead to drug-seeking behaviour. Certain drugs. especially ADH ANTIDIURETIC HORMONE (vasopressin). OPIOID ANALGESICS. such as heroin. have a strong progression adhesion The mechanism by which cells form contacts into drug dependence. with one another and/or an appropriate substratum, e.g. abuse of drugs The nonmedical use of drugs. i.e. integrins, selectins. 'recreational use' without the intention to treat a disease. It is adhesion molecules See cell adhesion molecules. usually a pejorative term, reflecting how drugs can seriously ADI acceptable daily intake (environmental chemistry). interfere with health. See also misuse of drugs. adjuvant A chemical agent that augments the activity of accessory cells Cells that. -

Clinical Evaluation of a Complement Fixation Test Using Human Hydatid Cyst Fluid Antigen for the Detection of Echinococcus Granulosus Antibody
©Österr. Ges. f. Tropenmedizin u. Parasitologie, download unter www.biologiezentrum.at Mitt. Österr. Ges. Tropenmed. Parasitol. 14 (1992) 117-128 Section of Clinical Microbiology, Mayo Clinic and Mayo Foundation, Rochester, MN, USA (Head: J. P. Anhalt, Ph. D, M. D.) Clinical evaluation of a complement fixation test using human hydatid cyst fluid antigen for the detection of Echinococcus granulosus antibody F. Allerberger, G. D. Roberts, J. E. Rosenblatt Introduction Complement fixation (CF) is a commonly used serodiagnostic test for hydatid disease, especially recommended for the evaluation of cure after surgery or for checking on treatment with antihelminthic drugs (9). However, the absence of an FDA-approved standardized antigen makes it difficult to assess the performance of different CF tests by using published studies. The objectives of this study were to determine sensitivity, specificity, negative predictive value and positive predictive value of a CF test using human hydatid cyst fluid as antigen and to evaluate the value of this test presently used at the Mayo Clinic in following the progress of treatment. Methods Complement-fixation test A micro-volume complement-fixation (CF) test was performed according to a technique described elsewhere (26). Briefly, the sera under examination were inactivated at 56° C for 30 min. in a waterbath. Using a dropper pipette 0.025 ml serum, 0.05 ml comple- ment (5C'H50) and 0.025 ml antigen were placed in U-shaped microtiter plates. Veronal buffer saline was used as diluent. After the plates had stood overnight at 4° C, 0.025 ml hemolysin with 0.28% sheep erythrocytes was added to each cup. -

Laboratory Manual Basic Immunology Edited By
Faculty of Science Faculty of Science Laboratory Manual Laboratory Manual Basic Immunology Bachelor of Biotechnology (Hons.) Bachelor of Biotechnology (Hons.) Copyright © 2018 Lincoln University College, Malaysia All rights reserved No part of this book can be reproduced or transmitted by any means, electronic or mechanical, including photocopying recording or by any information storage and retrieval system without prior written permission from the publisher. Edited By: Dr. Idris Adewale Ahmed LINCOLN UNIVERSITY COLLEGE Wisma Lincoln, No. 12,14,16 & 18, Jalan SS 6/12, Off Jalan Perbandaran 47301 Petaling, Jaya, Selangor Darul Ehsan, Malaysia Tel.: +603-7806 3478 Fax: +603-7806 3479 Toll Free: 1-300-880-111 E-mail: [email protected] [email protected] Web: www.lucp.net www.lincoln.edu.my ISBN: 978-967-2257-04-2 Basic Immunology Table of Content Experiments: Page 1 Laboratory safety rules 1 2 Agglutination reaction 6 3 Complement fixation test 9 4 Radial immuno-diffusion techniques 12 5 Dot enzyme linked immunosorbent assay (Dot ELISA) 15 LINCOLN UNIVERSITY COLLEGE FACULTY OF SCIENCE (DEPARTMENT OF BIOTECHNOLOGY) LABORATORY SAFETY RULES The following rules must be obeyed by all students in the science laboratory of the faculty. Wilful or repeated in advertent non-compliance may result in dismissal or suspension from the laboratories • No entry without permission: - Outsiders are not allowed to enter the laboratory without permission. - No student is allowed to enter the laboratory unless permission has been given by a laboratory assistant or a lecturer. • At work in the laboratory: - No experiment may be attempted without the knowledge and permission of a lecturer. -

Kuby Immunology
Immunology – II BC402 Immunological Methods and Applications ASHUTOSH SINGH Disclaimer: University of Lucknow does not subscribe to unauthorised usage of copyrighted contents from books, journals, other publications either in soft or hard copy of any nature including internet related contents. However, fair use at limited level of copyrighted material such as for commentary, criticism, news reporting, research and teaching as well as e-content of lectures, presentations and teaching related material may be incorporated in these e-teaching files. No commercial usage is allowed for all such material by the University and is authorised only for teaching purposes with all sanctity, these are added on our university web portal. Suggested Reading: Immunology by Barbara Anne Osborne and Janis Kuby Terminologies In vivo Involve whole animal In vitro Defined populations of immune cells are studied under controlled lab conditions Study of immune system requires suitable animal models For vaccine development – is the animal model susceptible to the disease? Mouse most often used Inbred strains reduce variation caused by differences in genetic backgrounds o 20 or more generations of brother-sister mating Have to abide by IACUC guidelines Adoptive Transfer Immune system of model animal can be eliminated Replaced with immune cells of animal to be studied Polyclonal Antibodies Are Secreted by Multiple Clones of Antigen-Specific B Cells Use of polyclonal antibodies Immunizing animal (mouse, rabbit) or human with antigen one or more times