Characterisation of Isomirs in Stem Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 ICR-Geneset Gene List
ICR-geneset Gene List. IMAGE ID UniGene Locus Name Cluster 20115 Hs.62185 SLC9A6 solute carrier family 9 (sodium/hydrogen exchanger), isoform 6 21738 21899 Hs.78353 SRPK2 SFRS protein kinase 2 21908 Hs.79133 CDH8 cadherin 8, type 2 22040 Hs.151738 MMP9 matrix metalloproteinase 9 (gelatinase B, 92kD gelatinase, 92kD type IV collagenase) 22411 Hs.183 FY Duffy blood group 22731 Hs.1787 PHRET1 PH domain containing protein in retina 1 22859 Hs.356487 ESTs 22883 Hs.150926 FPGT fucose-1-phosphate guanylyltransferase 22918 Hs.346868 EBNA1BP2 EBNA1 binding protein 2 23012 Hs.158205 BLZF1 basic leucine zipper nuclear factor 1 (JEM-1) 23073 Hs.284244 FGF2 fibroblast growth factor 2 (basic) 23173 Hs.151051 MAPK10 mitogen-activated protein kinase 10 23185 Hs.289114 TNC tenascin C (hexabrachion) 23282 Hs.8024 IK IK cytokine, down-regulator of HLA II 23353 23431 Hs.50421 RB1CC1 RB1-inducible coiled-coil 1 23514 23548 Hs.71848 Human clone 23548 mRNA sequence 23629 Hs.135587 Human clone 23629 mRNA sequence 23658 Hs.265855 SETMAR SET domain and mariner transposase fusion gene 23676 Hs.100841 Homo sapiens clone 23676 mRNA sequence 23772 Hs.78788 LZTR1 leucine-zipper-like transcriptional regulator, 1 23776 Hs.75438 QDPR quinoid dihydropteridine reductase 23804 Hs.343586 ZFP36 zinc finger protein 36, C3H type, homolog (mouse) 23831 Hs.155247 ALDOC aldolase C, fructose-bisphosphate 23878 Hs.99902 OPCML opioid binding protein/cell adhesion molecule-like 23903 Hs.12526 Homo sapiens clone 23903 mRNA sequence 23932 Hs.368063 Human clone 23932 mRNA sequence 24004 -

Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence After Radical Prostatectomy
Int. J. Mol. Sci. 2015, 16, 3856-3869; doi:10.3390/ijms16023856 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence after Radical Prostatectomy Chinyere Ibeawuchi 1, Hartmut Schmidt 2, Reinhard Voss 3, Ulf Titze 4, Mahmoud Abbas 5, Joerg Neumann 6, Elke Eltze 7, Agnes Marije Hoogland 8, Guido Jenster 9, Burkhard Brandt 10 and Axel Semjonow 1,* 1 Prostate Center, Department of Urology, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 2 Center for Laboratory Medicine, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 3 Interdisciplinary Center for Clinical Research, University of Muenster, Albert-Schweitzer-Campus 1, Gebaeude D3, Domagkstrasse 3, Muenster D-48149, Germany; E-Mail: [email protected] 4 Pathology, Lippe Hospital Detmold, Röntgenstrasse 18, Detmold D-32756, Germany; E-Mail: [email protected] 5 Institute of Pathology, Mathias-Spital-Rheine, Frankenburg Street 31, Rheine D-48431, Germany; E-Mail: [email protected] 6 Institute of Pathology, Klinikum Osnabrueck, Am Finkenhuegel 1, Osnabrueck D-49076, Germany; E-Mail: [email protected] 7 Institute of Pathology, Saarbrücken-Rastpfuhl, Rheinstrasse 2, Saarbrücken D-66113, Germany; E-Mail: [email protected] 8 Department -

Download The
PROBING THE INTERACTION OF ASPERGILLUS FUMIGATUS CONIDIA AND HUMAN AIRWAY EPITHELIAL CELLS BY TRANSCRIPTIONAL PROFILING IN BOTH SPECIES by POL GOMEZ B.Sc., The University of British Columbia, 2002 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Experimental Medicine) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2010 © Pol Gomez, 2010 ABSTRACT The cells of the airway epithelium play critical roles in host defense to inhaled irritants, and in asthma pathogenesis. These cells are constantly exposed to environmental factors, including the conidia of the ubiquitous mould Aspergillus fumigatus, which are small enough to reach the alveoli. A. fumigatus is associated with a spectrum of diseases ranging from asthma and allergic bronchopulmonary aspergillosis to aspergilloma and invasive aspergillosis. Airway epithelial cells have been shown to internalize A. fumigatus conidia in vitro, but the implications of this process for pathogenesis remain unclear. We have developed a cell culture model for this interaction using the human bronchial epithelium cell line 16HBE and a transgenic A. fumigatus strain expressing green fluorescent protein (GFP). Immunofluorescent staining and nystatin protection assays indicated that cells internalized upwards of 50% of bound conidia. Using fluorescence-activated cell sorting (FACS), cells directly interacting with conidia and cells not associated with any conidia were sorted into separate samples, with an overall accuracy of 75%. Genome-wide transcriptional profiling using microarrays revealed significant responses of 16HBE cells and conidia to each other. Significant changes in gene expression were identified between cells and conidia incubated alone versus together, as well as between GFP positive and negative sorted cells. -

Ten Commandments for a Good Scientist
Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Wageningen 2010 Thesis committee Thesis supervisors Prof. dr. ir. Ivonne M.C.M. Rietjens Professor of Toxicology Wageningen University Prof. dr. Albertinka J. Murk Personal chair at the sub-department of Toxicology Wageningen University Thesis co-supervisor Dr. ir. Jacques J.M. Vervoort Associate professor at the Laboratory of Biochemistry Wageningen University Other members Prof. dr. Michael R. Muller, Wageningen University Prof. dr. ir. Huub F.J. Savelkoul, Wageningen University Prof. dr. Everardus J. van Zoelen, Radboud University Nijmegen Dr. ir. Toine F.H. Bovee, RIKILT, Wageningen This research was conducted under the auspices of the Graduate School VLAG Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Thesis submitted in fulfillment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Tuesday 14 September 2010 at 4 p.m. in the Aula Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds. Ana María Sotoca Covaleda Thesis Wageningen University, Wageningen, The Netherlands, 2010, With references, and with summary in Dutch. ISBN: 978-90-8585-707-5 “Caminante no hay camino, se hace camino al andar. Al andar se hace camino, y al volver la vista atrás se ve la senda que nunca se ha de volver a pisar” - Antonio Machado – A mi madre. -

Predicting Clinical Response to Treatment with a Soluble Tnf-Antagonist Or Tnf, Or a Tnf Receptor Agonist
(19) TZZ _ __T (11) EP 2 192 197 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 02.06.2010 Bulletin 2010/22 C12Q 1/68 (2006.01) (21) Application number: 08170119.5 (22) Date of filing: 27.11.2008 (84) Designated Contracting States: (72) Inventor: The designation of the inventor has not AT BE BG CH CY CZ DE DK EE ES FI FR GB GR yet been filed HR HU IE IS IT LI LT LU LV MC MT NL NO PL PT RO SE SI SK TR (74) Representative: Habets, Winand Designated Extension States: Life Science Patents AL BA MK RS PO Box 5096 6130 PB Sittard (NL) (71) Applicant: Vereniging voor Christelijk Hoger Onderwijs, Wetenschappelijk Onderzoek en Patiëntenzorg 1081 HV Amsterdam (NL) (54) Predicting clinical response to treatment with a soluble tnf-antagonist or tnf, or a tnf receptor agonist (57) The invention relates to methods for predicting a clinical response to a therapy with a soluble TNF antagonist, TNF or a TNF receptor agonist and a kit for use in said methods. EP 2 192 197 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 192 197 A1 Description [0001] The invention relates to methods for predicting a clinical response to a treatment with a soluble TNF antagonist, with TNF or a TNF receptor agonist using expression levels of genes of the Type I INF pathway and a kit for use in said 5 methods. In another aspect, the invention relates to a method for evaluating a pharmacological effect of a treatment with a soluble TNF antagonist, TNF or a TNF receptor agonist. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

A Dissertation Entitled the Androgen Receptor
A Dissertation entitled The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit Submitted to the Graduate Faculty as partial fulfillment of the requirements for the PhD Degree in Biomedical science Dr. Manohar Ratnam, Committee Chair Dr. Lirim Shemshedini, Committee Member Dr. Robert Trumbly, Committee Member Dr. Edwin Sanchez, Committee Member Dr. Beata Lecka -Czernik, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2011 Copyright 2011, Mesfin Gonit This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit As partial fulfillment of the requirements for the PhD Degree in Biomedical science The University of Toledo August 2011 Prostate cancer depends on the androgen receptor (AR) for growth and survival even in the absence of androgen. In the classical models of gene activation by AR, ligand activated AR signals through binding to the androgen response elements (AREs) in the target gene promoter/enhancer. In the present study the role of AREs in the androgen- independent transcriptional signaling was investigated using LP50 cells, derived from parental LNCaP cells through extended passage in vitro. LP50 cells reflected the signature gene overexpression profile of advanced clinical prostate tumors. The growth of LP50 cells was profoundly dependent on nuclear localized AR but was independent of androgen. Nevertheless, in these cells AR was unable to bind to AREs in the absence of androgen. -

Antibodies Products
Chapter 2 : Gentaur Products List • Human Signal peptidase complex catalytic subunit • Human Sjoegren syndrome nuclear autoantigen 1 SSNA1 • Human Small proline rich protein 2A SPRR2A ELISA kit SEC11A SEC11A ELISA kit SpeciesHuman ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex catalytic subunit • Human Sjoegren syndrome scleroderma autoantigen 1 • Human Small proline rich protein 2B SPRR2B ELISA kit SEC11C SEC11C ELISA kit SpeciesHuman SSSCA1 ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 1 SPCS1 ELISA • Human Ski oncogene SKI ELISA kit SpeciesHuman • Human Small proline rich protein 2D SPRR2D ELISA kit kit SpeciesHuman • Human Ski like protein SKIL ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 2 SPCS2 ELISA • Human Skin specific protein 32 C1orf68 ELISA kit • Human Small proline rich protein 2E SPRR2E ELISA kit kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 3 SPCS3 ELISA • Human SLAIN motif containing protein 1 SLAIN1 ELISA kit • Human Small proline rich protein 2F SPRR2F ELISA kit kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptide CUB and EGF like domain • Human SLAIN motif containing protein 2 SLAIN2 ELISA kit • Human Small proline rich protein 2G SPRR2G ELISA kit containing protein 2 SCUBE2 ELISA kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptide CUB and EGF like domain • Human SLAM family member 5 CD84 ELISA kit • Human Small proline rich protein 3 SPRR3 ELISA kit containing protein -

Delineation of Key Regulatory Elements Identifies Points Of
DELINEATION OF KEY REGULATORY ELEMENTS IDENTIFIES POINTS OF VULNERABILITY IN THE MITOGEN-ACTIVATED SIGNALING NETWORK SUPPLEMENTARY MATERIALS List of contents Supplementary Figures with legends 1. Figure S1: Distribution of primary siRNA screen data, and standardization of assay procedure. 2. Figure S2: Scatter plot of screen data. 3. Figure S3: Functional relevance of the identified targets and Calculation of residence time from PDT and cell cycle distribution. 4. Figure S4: FACS profiles for ABL1 and AKT1. Table for data in Figure 5B. 5. Figure S5: Venn diagram showing the results of the comparative analysis of other screen results 6. Figure S6: Dose response profiles for the AKT1 + ABL1 inhibitor combination for CH1, list of the 14 cell lines and their description, effect of ABL1+AKT1 inhibitor combination on increase in apoptotic cells and G1 arrest in 14 cell lines, effects of CHEK1 inhibitor on combination C1,C2 on 4 cell lines. Supplementary Tables 1. Table S1: siRNA screen results for targeted kinases and phosphatases. 2. Table S2: Gene expression status of the validated hits. 3. Table S3: Role played by identified RNAi hits in regulation of cell cycle, the effect on PDTs along with phase-specific RTs. 4. Table S4: List of molecules classified as cell cycle targets. 5. Table S5: High confidence network used for graph theory analysis. 6. Table S6: Occurrences of nodes in shortest path networks. 7. Table S7: Network file used as SNAVI background. 8. Table S8: Classification of nodes present in modules according to specificity. Legends for tables Supplementary Experimental Procedures References Figure S1 A 450 400 G1 S 350 G2 300 250 200 150 100 50 Distribution of molecules Distribution 0 -6-4-20246 Z-score 350 200 400 G1 S 300 G2 150 300 250 200 100 200 150 100 50 100 Distribution of molecules 50 0 0 0 -4 -2 0 2 4 -4-20246 -4-20246 Z-score B PLK1 GAPDH PLCg BTK PLCg CDC2A PLCg CHEK1 PLCg MET Distribution profiles of complete primary screen and western blots showing knockdown efficiency. -
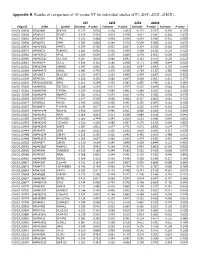
Appendix B. Results of Comparison of AI Versus NT for Individual Studies (D7e, D25e, D25x, D280x)
Appendix B. Results of comparison of AI versus NT for individual studies (d7E, d25E, d25X, d280X). d7E d25E d25X d280X Oligo ID cDNA Symbol Estimate P-value Estimate P-value Estimate P-value Estimate P-value OLIGO_00038 BF042860 RNF166 -0.123 0.634 0.194 0.469 -0.171 0.537 -0.302 0.263 OLIGO_00040 BF045167 POMT1 0.019 0.903 0.010 0.978 0.411 0.181 -0.204 0.476 OLIGO_00043 BF043567 ETFA 0.131 0.484 -0.094 0.706 -0.053 0.745 -0.331 0.056 OLIGO_00068 BF042951 GPX3 0.023 0.840 -0.249 0.370 -0.290 0.088 0.181 0.437 OLIGO_00070 AW464501 HPRT1 -0.079 0.785 0.055 0.837 0.224 0.259 -0.220 0.097 OLIGO_00074 BF040652 PLEKHC1 0.085 0.691 -0.256 0.399 0.309 0.156 0.133 0.423 OLIGO_00081 AW463422 PPIL1 0.109 0.600 0.142 0.686 0.379 0.061 -0.609 0.003 OLIGO_00086 AW462335 SULT1A1 -0.027 0.870 -0.083 0.845 0.123 0.675 0.520 0.121 OLIGO_00087 BF046074 CCL15 -0.306 0.132 -0.585 0.038 -0.771 0.038 0.844 0.020 OLIGO_00161 BM362588 USMG5 0.047 0.836 -0.136 0.628 0.547 0.043 0.279 0.064 OLIGO_00166 AW464473 UFD1L 0.083 0.742 0.372 0.538 0.759 0.132 0.285 0.418 OLIGO_00196 BF039617 BLOC1S2 0.011 0.972 0.153 0.699 0.424 0.187 0.030 0.931 OLIGO_00209 BF045356 GRB2 0.226 0.156 0.685 0.007 -0.008 0.961 0.212 0.177 OLIGO_00220 BP230002B20E6 NFX1 -0.161 0.294 -0.133 0.583 0.055 0.746 -0.211 0.290 OLIGO_00228 AW465641 TBC1D12 -0.248 0.236 -0.013 0.975 -0.223 0.496 0.044 0.821 OLIGO_00261 AW463949 TATDN1 0.020 0.914 -0.089 0.861 -0.248 0.611 -0.412 0.264 OLIGO_00308 AW464976 SNAPC3 0.021 0.911 -0.233 0.417 -0.054 0.782 0.004 0.979 OLIGO_00329 BF045997 PTP4A1 0.247 0.250 -
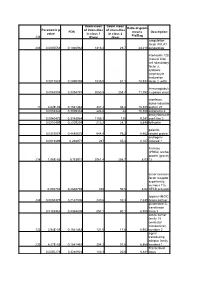
238 Parametric P- Value FDR Geom Mean of Intensities in Class 1 (Plate
Geom mean Geom mean Ratio of geom Parametric p- of intensities of intensities FDR means Description value in class 1 in class 2 Pla/Bag 238 (Plate) (Bag) coagulation factor XIII, A1 206 0,0005558 0,1940962 1313,3 29,7 44,219 polypeptide interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation 0,0017883 0,2490398 1535,5 81,1 18,933 factor 2, p40) Immunoglobuli 0,0063804 0,3394787 3050,5 258,7 11,792 n epsilon chain interferon, alpha-inducible 11 4,62E-05 0,1541482 347,2 33,4 10,395 protein 27 0,0035346 0,3039332 226,8 22 10,309 astrotactin 2 deoxyribonucle 0,0040473 0,3148984 1155,1 125 9,241 ase I-like 3 0,0010495 0,2205294 213,2 24,1 8,846 follistatin galectin- 0,0315078 0,4885675 644,8 76,2 8,462 related protein androgen- 0,0013899 0,238971 287 35,4 8,107 induced 1 A kinase (PRKA) anchor protein (gravin) 236 1,06E-05 0,115911 2061,4 256,7 8,03 12 tumor necrosis factor receptor superfamily, member 11a, 0,002746 0,2846759 405 50,5 8,02 NFKB activator lipoma HMGIC 248 0,0008375 0,2147006 240,6 32,3 7,449 fusion partner glutathione S- transferase 0,0183462 0,4368809 557,1 80,1 6,955 theta 1 solute carrier family 18 (vesicular monoamine), 122 3,94E-05 0,1541482 121,5 17,6 6,903 member 2 signal transducing adaptor family 233 6,27E-05 0,1541482 354,3 51,6 6,866 member 1 Transcribed 0,0050276 0,3240923 140,3 20,6 6,811 locus aldehyde dehydrogenas e 5 family, member A1 (succinate- semialdehyde dehydrogenas 0,0294865 0,4819829 236,6 38,2 6,194 e) guanylate cyclase activator 1A 0,0074099 0,3504479 339,1 55 6,165 -

Supplemental Table 1A. Differential Gene Expression Profile of Adehcd40l and Adehnull Treated Cells Vs Untreated Cells
Supplemental Table 1a. Differential Gene Expression Profile of AdEHCD40L and AdEHNull treated cells vs Untreated Cells Fold change Regulation Fold change Regulation ([AdEHCD40L] vs ([AdEHCD40L] ([AdEHNull] vs ([AdEHNull] vs Probe Set ID [Untreated]) vs [Untreated]) [Untreated]) [Untreated]) Gene Symbol Gene Title RefSeq Transcript ID NM_001039468 /// NM_001039469 /// NM_004954 /// 203942_s_at 2.02 down 1.00 down MARK2 MAP/microtubule affinity-regulating kinase 2 NM_017490 217985_s_at 2.09 down 1.00 down BAZ1A fibroblastbromodomain growth adjacent factor receptorto zinc finger 2 (bacteria-expressed domain, 1A kinase, keratinocyte NM_013448 /// NM_182648 growth factor receptor, craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer 203638_s_at 2.10 down 1.01 down FGFR2 syndrome, Jackson-Weiss syndrome) NM_000141 /// NM_022970 1570445_a_at 2.07 down 1.01 down LOC643201 hypothetical protein LOC643201 XM_001716444 /// XM_001717933 /// XM_932161 231763_at 3.05 down 1.02 down POLR3A polymerase (RNA) III (DNA directed) polypeptide A, 155kDa NM_007055 1555368_x_at 2.08 down 1.04 down ZNF479 zinc finger protein 479 NM_033273 /// XM_001714591 /// XM_001719979 241627_x_at 2.15 down 1.05 down FLJ10357 hypothetical protein FLJ10357 NM_018071 223208_at 2.17 down 1.06 down KCTD10 potassium channel tetramerisation domain containing 10 NM_031954 219923_at 2.09 down 1.07 down TRIM45 tripartite motif-containing 45 NM_025188 242772_x_at 2.03 down 1.07 down Transcribed locus 233019_at 2.19 down 1.08 down CNOT7 CCR4-NOT transcription complex, subunit 7 NM_013354