Antibodies Products
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Information to Users
INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of tftis reproduction is dependent upon the quality of the copy sutHnitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. in the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with smal overlaps. Photographs included in the original manuscript have been reproduced xerographically in this copy. Higher quality 6" x 9* black and white photographic prints are available for any photographs or Uustrations appearing in this copy for an additional charge. Contact UMI directly to order. ProQuest Information and Learning 300 North Zeeb Road, Atm Arbor, Ml 48106-1346 USA 800-521-0600 UMT MOLECULAR ANALYSIS OF L7/PCP-2 MESSENGER RNA AND ITS INTERACTING PROTEINS DISSERTATION Presented in Partial Fulfillment of the Requirements for The Degree Doctor of Philosophy in Ohio State Biochemistry Program of The Ohio State University By Xulun Zhang, M.S. ***** The Ohio State University 2001 Dissertation Committee: Approved by Professor John D. -

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

Plant SET Domain-Containing Proteins: Structure, Function and Regulation
Biochimica et Biophysica Acta 1769 (2007) 316–329 www.elsevier.com/locate/bbaexp Review Plant SET domain-containing proteins: Structure, function and regulation Danny W-K Ng, Tao Wang, Mahesh B. Chandrasekharan 1, Rodolfo Aramayo, ⁎ Sunee Kertbundit 2, Timothy C. Hall Institute of Developmental and Molecular Biology and Department of Biology, Texas A&M University, College Station, TX 77843-3155, USA Received 27 October 2006; received in revised form 3 April 2007; accepted 4 April 2007 Available online 12 April 2007 Abstract Modification of the histone proteins that form the core around which chromosomal DNA is looped has profound epigenetic effects on the accessibility of the associated DNA for transcription, replication and repair. The SET domain is now recognized as generally having methyltransferase activity targeted to specific lysine residues of histone H3 or H4. There is considerable sequence conservation within the SET domain and within its flanking regions. Previous reviews have shown that SET proteins from Arabidopsis and maize fall into five classes according to their sequence and domain architectures. These classes generally reflect specificity for a particular substrate. SET proteins from rice were found to fall into similar groupings, strengthening the merit of the approach taken. Two additional classes, VI and VII, were established that include proteins with truncated/ interrupted SET domains. Diverse mechanisms are involved in shaping the function and regulation of SET proteins. These include protein–protein interactions through both intra- and inter-molecular associations that are important in plant developmental processes, such as flowering time control and embryogenesis. Alternative splicing that can result in the generation of two to several different transcript isoforms is now known to be widespread. -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

1 ICR-Geneset Gene List
ICR-geneset Gene List. IMAGE ID UniGene Locus Name Cluster 20115 Hs.62185 SLC9A6 solute carrier family 9 (sodium/hydrogen exchanger), isoform 6 21738 21899 Hs.78353 SRPK2 SFRS protein kinase 2 21908 Hs.79133 CDH8 cadherin 8, type 2 22040 Hs.151738 MMP9 matrix metalloproteinase 9 (gelatinase B, 92kD gelatinase, 92kD type IV collagenase) 22411 Hs.183 FY Duffy blood group 22731 Hs.1787 PHRET1 PH domain containing protein in retina 1 22859 Hs.356487 ESTs 22883 Hs.150926 FPGT fucose-1-phosphate guanylyltransferase 22918 Hs.346868 EBNA1BP2 EBNA1 binding protein 2 23012 Hs.158205 BLZF1 basic leucine zipper nuclear factor 1 (JEM-1) 23073 Hs.284244 FGF2 fibroblast growth factor 2 (basic) 23173 Hs.151051 MAPK10 mitogen-activated protein kinase 10 23185 Hs.289114 TNC tenascin C (hexabrachion) 23282 Hs.8024 IK IK cytokine, down-regulator of HLA II 23353 23431 Hs.50421 RB1CC1 RB1-inducible coiled-coil 1 23514 23548 Hs.71848 Human clone 23548 mRNA sequence 23629 Hs.135587 Human clone 23629 mRNA sequence 23658 Hs.265855 SETMAR SET domain and mariner transposase fusion gene 23676 Hs.100841 Homo sapiens clone 23676 mRNA sequence 23772 Hs.78788 LZTR1 leucine-zipper-like transcriptional regulator, 1 23776 Hs.75438 QDPR quinoid dihydropteridine reductase 23804 Hs.343586 ZFP36 zinc finger protein 36, C3H type, homolog (mouse) 23831 Hs.155247 ALDOC aldolase C, fructose-bisphosphate 23878 Hs.99902 OPCML opioid binding protein/cell adhesion molecule-like 23903 Hs.12526 Homo sapiens clone 23903 mRNA sequence 23932 Hs.368063 Human clone 23932 mRNA sequence 24004 -

Download The
PROBING THE INTERACTION OF ASPERGILLUS FUMIGATUS CONIDIA AND HUMAN AIRWAY EPITHELIAL CELLS BY TRANSCRIPTIONAL PROFILING IN BOTH SPECIES by POL GOMEZ B.Sc., The University of British Columbia, 2002 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Experimental Medicine) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2010 © Pol Gomez, 2010 ABSTRACT The cells of the airway epithelium play critical roles in host defense to inhaled irritants, and in asthma pathogenesis. These cells are constantly exposed to environmental factors, including the conidia of the ubiquitous mould Aspergillus fumigatus, which are small enough to reach the alveoli. A. fumigatus is associated with a spectrum of diseases ranging from asthma and allergic bronchopulmonary aspergillosis to aspergilloma and invasive aspergillosis. Airway epithelial cells have been shown to internalize A. fumigatus conidia in vitro, but the implications of this process for pathogenesis remain unclear. We have developed a cell culture model for this interaction using the human bronchial epithelium cell line 16HBE and a transgenic A. fumigatus strain expressing green fluorescent protein (GFP). Immunofluorescent staining and nystatin protection assays indicated that cells internalized upwards of 50% of bound conidia. Using fluorescence-activated cell sorting (FACS), cells directly interacting with conidia and cells not associated with any conidia were sorted into separate samples, with an overall accuracy of 75%. Genome-wide transcriptional profiling using microarrays revealed significant responses of 16HBE cells and conidia to each other. Significant changes in gene expression were identified between cells and conidia incubated alone versus together, as well as between GFP positive and negative sorted cells. -

EFEMP1 Expression Promotes in Vivo Tumor Growth in Human Pancreatic Adenocarcinoma
Published OnlineFirst February 10, 2009; DOI: 10.1158/1541-7786.MCR-08-0132 EFEMP1 Expression Promotes In vivo Tumor Growth in Human Pancreatic Adenocarcinoma Hendrik Seeliger,1 Peter Camaj,1 Ivan Ischenko,1 Axel Kleespies,1 Enrico N. De Toni,2 Susanne E. Thieme,4 Helmut Blum,4 Gerald Assmann,3 Karl-Walter Jauch,1 and Christiane J. Bruns1 Departments of 1Surgery and 2Gastroenterology and 3Institute of Pathology, Munich University Medical Center; 4Gene Center, Munich University, Munich, Germany Abstract VEGF-driven angiogenesis and antiapoptotic mechanisms. The progression of pancreatic cancer is dependent on Hence, EFEMP1 is a promising candidate for assessing local tumor growth, angiogenesis, and metastasis. prognosis and individualizing therapy in a clinical tumor EFEMP1, a recently discovered member of the fibulin setting. (Mol Cancer Res 2009;7(2):189–98) family, was characterized with regard to these key elements of pancreatic cancer progression. Differential gene expression was assessed Introduction by mRNA microarray hybridization in FG human Pancreatic cancer is one of the leading causes of cancer- pancreatic adenocarcinoma cells and L3.6pl cells, related deaths in western countries. Despite improved multi- a highly metastatic variant of FG. In vivo orthotopic modal therapeutic regimens, its prognosis has improved only tumor growth of EFEMP1-transfected FG cells was marginally, resulting in a total 5-year survival rate, which is examined in nude mice. To assess the angiogenic still as low as 5% (1). More recently, agents targeted against properties of EFEMP1, vascular endothelial growth molecular determinants of cancer cells or tumor vessels, or factor (VEGF) production of tumor cells, both, have been tested successfully in clinical trials to expand endothelial cell proliferation and migration, and the therapeutic spectrum (2-4). -
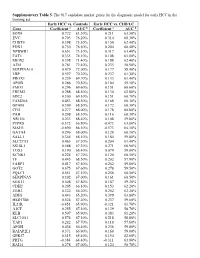
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

DM Domain Proteins in Sexual Regulation 875 Transformation Marker Prf4 (Containing the Mutant Rol-6 Allele Green fluorescent Protein (GFP) Coding Region
Development 126, 873-881 (1999) 873 Printed in Great Britain © The Company of Biologists Limited 1999 DEV5283 Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms Woelsung Yi1 and David Zarkower1,2,* 1Biochemistry, Molecular Biology and Biophysics Graduate Program, and 2Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, MN 55455, USA *Author for correspondence (e-mail: [email protected]) Accepted 17 December 1998; published on WWW 2 February 1999 SUMMARY Although most animals occur in two sexes, the molecular transcription, resulting in expression in both sexes, and the pathways they employ to control sexual development vary vitellogenin genes have potential MAB-3 binding sites considerably. The only known molecular similarity upstream of their transcriptional start sites. MAB-3 binds between phyla in sex determination is between two genes, to a site in the vit-2 promoter in vitro, and this site is mab-3 from C. elegans, and doublesex (dsx) from required in vivo to prevent transcription of a vit-2 reporter Drosophila. Both genes contain a DNA binding motif called construct in males, suggesting that MAB-3 is a direct a DM domain and they regulate similar aspects of sexual repressor of vitellogenin transcription. This is the first development, including yolk protein synthesis and direct link between the sex determination regulatory peripheral nervous system differentiation. Here we show pathway and sex-specific structural genes in C. elegans, and that MAB-3, like the DSX proteins, is a direct regulator of it suggests that nematodes and insects use at least some of yolk protein gene transcription. -

Computational Investigation of Thiol-Based Redox Modifications in Proteins: Redox-Active Disulfides, Zn2+ Sites and Their Associations
THESIS SUBMITTED IN TOTAL FULFILLMENT OF THE REQUIREMENTS OF THE DEGREE OF DOCTOR OF PHILOSOPHY Computational investigation of thiol‐based redox modifications in proteins: redox‐active disulfides, 2+ Zn sites and their associations Application of tools of bioinformatics in health and disease Dhakshinari Vihara Kumari Hulugalle March 2013 Victor Chang Cardiac Research Institute School of Medical Sciences, Faculty of Medicine, University of New South Wales PLEASE TYPE THE UNIVERSITY OF NEW SOUTH WALES Thesis/Dissertation Sheet Surname or Family name: HULUGALLE First name: DHAKSHINARI Other name/s: VIHARA KUMARI PhD Abbreviation for degree as given in the University calendar: School: SCHOOL OF MEDICAL SCIENCES Faculty: MEDICINE Title: Computational investigation of thiol-based redox modifications in proteins: redox-active disulfides, Zn2+ sites and their associations Abstract 350 words maximum: (PLEASE TYPE) Thiol based redox signalling is an emerging area of research in protein science. Reversible disulfide bonding and Zn2+ expulsion are two important but less explored oxidative thiol modifications associated with redox signalling. They have numerous implications in health and disease. Three computational methods have previously been developed to predict redox-active disulfides in proteins: redox pair protein (RP) method, ‘forbidden disulfides’ (FD) method and torsional energy (TE) method. These methods and other tools of bioinformatics are used in my study to investigate redox-active disulfides, Zn2+ sites and the association between FDs and Zn fingers in protein structures. The first objective of my study was to apply the RP method to predict novel proteins containing likely redox-active disulfides. Over 300 novel RP proteins were found during this study. Significant conformational changes associated with disulfide redox activity in RP protein structures were also identified. -

Predicting Clinical Response to Treatment with a Soluble Tnf-Antagonist Or Tnf, Or a Tnf Receptor Agonist
(19) TZZ _ __T (11) EP 2 192 197 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 02.06.2010 Bulletin 2010/22 C12Q 1/68 (2006.01) (21) Application number: 08170119.5 (22) Date of filing: 27.11.2008 (84) Designated Contracting States: (72) Inventor: The designation of the inventor has not AT BE BG CH CY CZ DE DK EE ES FI FR GB GR yet been filed HR HU IE IS IT LI LT LU LV MC MT NL NO PL PT RO SE SI SK TR (74) Representative: Habets, Winand Designated Extension States: Life Science Patents AL BA MK RS PO Box 5096 6130 PB Sittard (NL) (71) Applicant: Vereniging voor Christelijk Hoger Onderwijs, Wetenschappelijk Onderzoek en Patiëntenzorg 1081 HV Amsterdam (NL) (54) Predicting clinical response to treatment with a soluble tnf-antagonist or tnf, or a tnf receptor agonist (57) The invention relates to methods for predicting a clinical response to a therapy with a soluble TNF antagonist, TNF or a TNF receptor agonist and a kit for use in said methods. EP 2 192 197 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 192 197 A1 Description [0001] The invention relates to methods for predicting a clinical response to a treatment with a soluble TNF antagonist, with TNF or a TNF receptor agonist using expression levels of genes of the Type I INF pathway and a kit for use in said 5 methods. In another aspect, the invention relates to a method for evaluating a pharmacological effect of a treatment with a soluble TNF antagonist, TNF or a TNF receptor agonist. -

A W-Linked DM-Domain Gene, DM-W, Participates in Primary Ovary Development in Xenopus Laevis
A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis Shin Yoshimoto*, Ema Okada*, Hirohito Umemoto*, Kei Tamura*, Yoshinobu Uno†, Chizuko Nishida-Umehara†, Yoichi Matsuda†, Nobuhiko Takamatsu*, Tadayoshi Shiba*, and Michihiko Ito*‡ *Department of Bioscience, School of Science, Kitasato University, 1-15-1 Kitasato, Sagamihara, Kanagawa 228-8555, Japan; and †Division of Genome Dynamics, Creative Research Initiative ‘‘Sousei,’’ Hokkaido University, North10 West8, Kita-ku, Sapporo, Hokkaido 060-0810, Japan Communicated by Satoshi Omura,¯ The Kitasato Institute, Tokyo, Japan, December 27, 2007 (received for review September 11, 2007) In the XX/XY sex-determining system, the Y-linked SRY genes of In some species of amphibians, sex determination is controlled most mammals and the DMY/Dmrt1bY genes of the teleost fish genetically (13), even though the animals’ sex chromosomes are medaka have been characterized as sex-determining genes that morphologically indistinguishable from the autosomes. The trigger formation of the testis. However, the molecular mechanism South African clawed frog Xenopus laevis uses the ZZ/ZW of the ZZ/ZW-type system in vertebrates, including the clawed frog system, which was demonstrated by backcrosses between sex- Xenopus laevis, is unknown. Here, we isolated an X. laevis female reversed and normal individuals (14), but its sex chromosomes genome-specific DM-domain gene, DM-W, and obtained molecular have not yet been identified. Moreover, as with other animals evidence of a W-chromosome in this species. The DNA-binding that use the ZZ/ZW system, no sex-determining gene(s) has domain of DM-W showed a strikingly high identity (89%) with that been identified.