Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Loss of Mafb and Maf Distorts Myeloid Cell Ratios and Disrupts Fetal Mouse Testis Vascularization and Organogenesisǂ
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441488; this version posted April 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Loss of Mafb and Maf distorts myeloid cell ratios and disrupts fetal mouse testis vascularization and organogenesisǂ 5 Shu-Yun Li1,5, Xiaowei Gu1,5, Anna Heinrich1, Emily G. Hurley1,2,3, Blanche Capel4, and Tony DeFalco1,2* 1Division of Reproductive Sciences, Cincinnati Children’s Hospital Medical Center, Cincinnati, 10 OH 45229, USA 2Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH 45267 USA 3Department of Obstetrics and Gynecology, University of Cincinnati College of Medicine, Cincinnati, OH 45267 USA 15 4Department of Cell Biology, Duke University Medical Center, Durham, NC 27710 USA 5These authors contributed equally to this work. ǂThis work was supported by the National Institutes of Health (R37HD039963 to BC, R35GM119458 to TD, R01HD094698 to TD, F32HD058433 to TD); March of Dimes (1-FY10- 355 to BC, Basil O’Connor Starter Scholar Award 5-FY14-32 to TD); Lalor Foundation 20 (postdoctoral fellowship to SL); and Cincinnati Children’s Hospital Medical Center (Research Innovation and Pilot funding, Trustee Award, and developmental funds to TD). *Corresponding Author: Tony DeFalco E-mail: [email protected] 25 Address: Division of Reproductive Sciences Cincinnati Children’s Hospital Medical Center 3333 Burnet Avenue, MLC 7045 Cincinnati, OH 45229 USA Phone: +1-513-803-3988 30 Fax: +1-513-803-1160 bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441488; this version posted April 26, 2021. -

Tomo-Seq Identifies SOX9 As a Key Regulator of Cardiac Fibrosis During Ischemic Injury
myocardial myocardial Eva van ◼ osis fibr SOX9 transcription ◼ PhD* PhD PhD PhD MSc, PhD naarden, MSc, PhD naarden, PhD* ventricular remodeling Correspondence to: Correspondence Rooij, MSc, PhD, Hubrecht Department of Institute, KNAW University Medical Cardiology, Uppsalalaan 8, Center Utrecht, The Netherlands. 3584CT Utrecht, E-mail [email protected] of Funding, see page 1408 Sources Key Words: ischemia ◼ © 2017 American Heart Association, Inc. *Drs. Lacraz and Junker contributed equally. Grégory P.A. Lacraz, MSc, P.A. Grégory MSc, Jan Philipp Junker, Monika M. Gladka, MSc, MSc Bas Molenaar, Scholman, MSc Koen T. MSc Marta Vigil-Garcia, BS Danielle Versteeg, BS Hesther de Ruiter, MSc, Vermunt, Marit W. MSc, Creyghton, Menno P. Manon M.H. Huibers, Nicolaas de Jonge, MD Alexander van Oude- Eva van Rooij, MSc, PhD 2017;136:1396–1409. DOI: 10.1161/CIRCULATIONAHA.117.027832 DOI: 2017;136:1396–1409. Circulation. blunted the cardiac fibrotic fibrotic blunted the cardiac Sox9 ). Subsequent correlation analysis allowed). Subsequent correlation Serca2 Editorial, see p 1410 , and Nppa Based on the exact local expression cues, tomo-seq can Based on the exact local expression Cardiac ischemic injury induces a pathological remodeling ischemic injury induces a pathological remodeling Cardiac , Although genome-wide transcriptome analysis on diseased tissues Tracing transcriptional differences with a high spatial resolution with a high spatial resolution transcriptional differences Tracing Col1a2 October 10, 2017 October 10, 1396 CONCLUSIONS: novel genes and key transcription factors involved in specific serve to reveal able to unveil the Using tomo-seq, we were remodeling. aspects of cardiac pointing fibrosis, of cardiac of SOX9 as a key regulator unknown relevance fibrosis. -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

The Versatile Functions of Sox9 in Development, Stem Cells, And
Genes & Diseases (2014) 1, 149e161 HOSTED BY Available online at www.sciencedirect.com ScienceDirect journal homepage: http://ees.elsevier.com/gendis/default.asp REVIEW ARTICLE The versatile functions of Sox9 in development, stem cells, and human diseases Alice Jo a, Sahitya Denduluri a, Bosi Zhang a, Zhongliang Wang a,b, Liangjun Yin a,b, Zhengjian Yan a,b, Richard Kang a, Lewis L. Shi a, James Mok a, Michael J. Lee a, Rex C. Haydon a,* a Molecular Oncology Laboratory, Department of Orthopaedic Surgery and Rehabilitation Medicine, The University of Chicago Medical Center, Chicago, IL 60637, USA b Departments of Orthopaedic Surgery, The Affiliated Hospitals of Chongqing Medical University, Chongqing 400046, China Received 3 September 2014; accepted 6 September 2014 Available online 16 October 2014 KEYWORDS Abstract The transcription factor Sox9 was first discoveredinpatientswithcampomelic Development; dysplasia, a haploinsufficiency disorder with skeletal deformities caused by dysregulation of Sox9; Sox9 expression during chondrogenesis. Since then, its role as a cell fate determiner during Stem cells; embryonic development has been well characterized; Sox9 expression differentiates cells Transcription factor derived from all three germ layers into a large variety of specialized tissues and organs. How- ever, recent data has shown that ectoderm- and endoderm-derived tissues continue to express Sox9 in mature organs and stem cell pools, suggesting its role in cell maintenance and speci- fication during adult life. The versatility of Sox9 may be explained by a combination of post- transcriptional modifications, binding partners, and the tissue type in which it is expressed. Considering its importance during both development and adult life, it follows that dysregula- tion of Sox9 has been implicated in various congenital and acquired diseases, including fibrosis and cancer. -

Curcumin Attenuates Environment-Derived Osteoarthritis by Sox9/NF-Kb Signaling Axis
International Journal of Molecular Sciences Article Curcumin Attenuates Environment-Derived Osteoarthritis by Sox9/NF-kB Signaling Axis Constanze Buhrmann 1,2, Aranka Brockmueller 1, Anna-Lena Mueller 1, Parviz Shayan 3 and Mehdi Shakibaei 1,* 1 Musculoskeletal Research Group and Tumor Biology, Chair of Vegetative Anatomy, Institute of Anatomy, Faculty of Medicine, Ludwig-Maximilian-University Munich, Pettenkoferstr. 11, D-80336 Munich, Germany; [email protected] (C.B.); [email protected] (A.B.); [email protected] (A.-L.M.) 2 Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Augsburg, Universitaetsstr. 2, D-86159 Augsburg, Germany 3 Department of Parasitology, Faculty of Veterinary Medicine, University of Tehran, Tehran 141556453, Iran; [email protected] * Correspondence: [email protected]; Tel.: +49-89-2180-72624 Abstract: Inflammation has a fundamental impact on the pathophysiology of osteoarthritis (OA), a common form of degenerative arthritis. It has previously been established that curcumin, a component of turmeric (Curcuma longa), has anti-inflammatory properties. This research evaluates the potentials of curcumin on the pathophysiology of OA in vitro. To explore the anti-inflammatory efficacy of curcumin in an inflamed joint, an osteoarthritic environment (OA-EN) model consisting of fibroblasts, T-lymphocytes, 3D-chondrocytes is constructed and co-incubated with TNF-α, antisense oligonucleotides targeting NF-kB (ASO-NF-kB), or an IkB-kinase (IKK) inhibitor (BMS-345541). Our results show that OA-EN, similar to TNF-α, suppresses chondrocyte viability, which is accompanied Citation: Buhrmann, C.; by a significant decrease in cartilage-specific proteins (collagen II, CSPG, Sox9) and an increase in NF- Brockmueller, A.; Mueller, A.-L.; kB-driven gene proteins participating in inflammation, apoptosis, and breakdown (NF-kB, MMP-9, Shayan, P.; Shakibaei, M. -
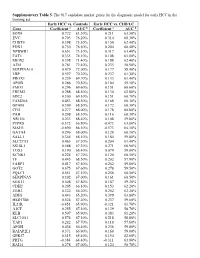
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

SUPPLEMENTARY MATERIAL Bone Morphogenetic Protein 4 Promotes
www.intjdevbiol.com doi: 10.1387/ijdb.160040mk SUPPLEMENTARY MATERIAL corresponding to: Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells SUMIYO MIMURA, MIKA SUGA, KAORI OKADA, MASAKI KINEHARA, HIROKI NIKAWA and MIHO K. FURUE* *Address correspondence to: Miho Kusuda Furue. Laboratory of Stem Cell Cultures, National Institutes of Biomedical Innovation, Health and Nutrition, 7-6-8, Saito-Asagi, Ibaraki, Osaka 567-0085, Japan. Tel: 81-72-641-9819. Fax: 81-72-641-9812. E-mail: [email protected] Full text for this paper is available at: http://dx.doi.org/10.1387/ijdb.160040mk TABLE S1 PRIMER LIST FOR QRT-PCR Gene forward reverse AP2α AATTTCTCAACCGACAACATT ATCTGTTTTGTAGCCAGGAGC CDX2 CTGGAGCTGGAGAAGGAGTTTC ATTTTAACCTGCCTCTCAGAGAGC DLX1 AGTTTGCAGTTGCAGGCTTT CCCTGCTTCATCAGCTTCTT FOXD3 CAGCGGTTCGGCGGGAGG TGAGTGAGAGGTTGTGGCGGATG GAPDH CAAAGTTGTCATGGATGACC CCATGGAGAAGGCTGGGG MSX1 GGATCAGACTTCGGAGAGTGAACT GCCTTCCCTTTAACCCTCACA NANOG TGAACCTCAGCTACAAACAG TGGTGGTAGGAAGAGTAAAG OCT4 GACAGGGGGAGGGGAGGAGCTAGG CTTCCCTCCAACCAGTTGCCCCAAA PAX3 TTGCAATGGCCTCTCAC AGGGGAGAGCGCGTAATC PAX6 GTCCATCTTTGCTTGGGAAA TAGCCAGGTTGCGAAGAACT p75 TCATCCCTGTCTATTGCTCCA TGTTCTGCTTGCAGCTGTTC SOX9 AATGGAGCAGCGAAATCAAC CAGAGAGATTTAGCACACTGATC SOX10 GACCAGTACCCGCACCTG CGCTTGTCACTTTCGTTCAG Suppl. Fig. S1. Comparison of the gene expression profiles of the ES cells and the cells induced by NC and NC-B condition. Scatter plots compares the normalized expression of every gene on the array (refer to Table S3). The central line -

Table S2.Up Or Down Regulated Genes in Tcof1 Knockdown Neuroblastoma N1E-115 Cells Involved in Differentbiological Process Anal
Table S2.Up or down regulated genes in Tcof1 knockdown neuroblastoma N1E-115 cells involved in differentbiological process analysed by DAVID database Pop Pop Fold Term PValue Genes Bonferroni Benjamini FDR Hits Total Enrichment GO:0044257~cellular protein catabolic 2.77E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 537 13588 1.944851 8.64E-07 8.64E-07 5.02E-07 process ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, USP17L5, FBXO11, RAD23B, NEDD8, UBE2V2, RFFL, CDC GO:0051603~proteolysis involved in 4.52E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 534 13588 1.93519 1.41E-06 7.04E-07 8.18E-07 cellular protein catabolic process ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, USP17L5, FBXO11, RAD23B, NEDD8, UBE2V2, RFFL, CDC GO:0044265~cellular macromolecule 6.09E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 609 13588 1.859332 1.90E-06 6.32E-07 1.10E-06 catabolic process ISG15, RBM8A, ATG7, LOC100046898, PSENEN, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, XRN2, USP17L5, FBXO11, RAD23B, UBE2V2, NED GO:0030163~protein catabolic process 1.81E-09 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 556 13588 1.87839 5.64E-06 1.41E-06 3.27E-06 ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice. -

Modeling Enhancer-Promoter Interactions with Attention-Based Neural Networks
bioRxiv preprint doi: https://doi.org/10.1101/219667; this version posted November 14, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Article Modeling Enhancer-Promoter Interactions with Attention-Based Neural Networks Weiguang Mao 1,2, Dennis Kostka 1,2,3 and Maria Chikina 1,2* 1 Department of Computational and Systems Biology, School of Medicine, University of Pittsburgh 2 Joint Carnegie Mellon - University of Pittsburgh Ph.D. Program in Computational Biology 3 Department of Developmental Biology, School of Medicine, University of Pittsburgh * Correspondence: [email protected] Academic Editor: name Version November 9, 2017 submitted to Cells 1 Abstract: Background: Gene regulatory sequences play critical roles in ensuring tightly controlled 2 RNA expression patterns that are essential in a large variety of biological processes. Specifically, 3 enhancer sequences drive expression of their target genes, and the availability of genome-wide 4 maps of enhancer-promoter interactions has opened up the possibility to use machine learning 5 approaches to extract and interpret features that define these interactions in different biological 6 contexts. Methods: Inspired by machine translation models we develop an attention-based neural 7 network model, EPIANN, to predict enhancer-promoter interactions based on DNA sequences. Codes 8 and data are available at https://github.com/wgmao/EPIANN. Results: Our approach accurately 9 predicts enhancer-promoter interactions across six cell lines. In addition, our method generates 10 pairwise attention scores at the sequence level, which specify how short regions in the enhancer and 11 promoter pair-up to drive the interaction prediction. -

Upregulation of the Transcription Factor TFAP2D Is Associated With
Fraune et al. Molecular Medicine (2020) 26:24 Molecular Medicine https://doi.org/10.1186/s10020-020-00148-4 RESEARCH ARTICLE Open Access Upregulation of the transcription factor TFAP2D is associated with aggressive tumor phenotype in prostate cancer lacking the TMPRSS2:ERG fusion Christoph Fraune1†, Luisa Harms1†, Franziska Büscheck1, Doris Höflmayer1, Maria Christina Tsourlakis1, Till S. Clauditz1, Ronald Simon1* , Katharina Möller1, Andreas M. Luebke1, Christina Möller-Koop1, Stefan Steurer1, Claudia Hube-Magg1, Guido Sauter1, Sören Weidemann1, Patrick Lebok1, David Dum1, Simon Kind1, Sarah Minner1, Jakob R. Izbicki2, Thorsten Schlomm3, Hartwig Huland4, Hans Heinzer4, Eike Burandt1, Alexander Haese4, Markus Graefen4 and Cornelia Schroeder2 Abstract Background: TFAP2D is a transcription factor important for modulating gene expression in embryogenesis. Its expression and prognostic role in prostate cancer has not been evaluated. Methods: Therefore, a tissue microarray containing 17,747 prostate cancer specimens with associated pathological, clinical, and molecular data was analyzed by immunohistochemistry to assess the role of TFAP2D. Results: TFAP2D expression was typically increased in prostate cancer as compared to adjacent non-neoplastic glands. TFAP2D staining was considered negative in 24.3% and positive in 75.7% of 13,545 interpretable cancers. TFAP2D staining was significantly linked to advanced tumor stage, high classical and quantitative Gleason grade, lymph node metastasis, and a positive surgical margin (p ≤ 0.0045). TFAP2D positivity was more common in ERG fusion positive (88.7%) than in ERG negative cancers (66.8%; p < 0.0001). Subset analyses in 3776 cancers with and 4722 cancers without TMPRSS2:ERG fusion revealed that associations with tumor phenotype and patient outcome were largely driven by the subset of ERG negative tumors. -

Co-Regulation of Hormone Receptors, Neuropeptides, and Steroidogenic Enzymes 2 Across the Vertebrate Social Behavior Network 3 4 Brent M
bioRxiv preprint doi: https://doi.org/10.1101/435024; this version posted October 4, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Co-regulation of hormone receptors, neuropeptides, and steroidogenic enzymes 2 across the vertebrate social behavior network 3 4 Brent M. Horton1, T. Brandt Ryder2, Ignacio T. Moore3, Christopher N. 5 Balakrishnan4,* 6 1Millersville University, Department of Biology 7 2Smithsonian Conservation Biology Institute, Migratory Bird Center 8 3Virginia Tech, Department of Biological Sciences 9 4East Carolina University, Department of Biology 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 1 bioRxiv preprint doi: https://doi.org/10.1101/435024; this version posted October 4, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Running Title: Gene expression in the social behavior network 2 Keywords: dominance, systems biology, songbird, territoriality, genome 3 Corresponding Author: 4 Christopher Balakrishnan 5 East Carolina University 6 Department of Biology 7 Howell Science Complex 8 Greenville, NC, USA 27858 9 [email protected] 10 2 bioRxiv preprint doi: https://doi.org/10.1101/435024; this version posted October 4, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity.