Upregulation of the Transcription Factor TFAP2D Is Associated With
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

HOOK3 Is a Scaffold for the Opposite-Polarity Microtubule-Based
bioRxiv preprint doi: https://doi.org/10.1101/508887; this version posted December 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. HOOK3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein and KIF1C Agnieszka A. Kendrick1, William B. Redwine1,2†, Phuoc Tien Tran1‡, Laura Pontano Vaites2, Monika Dzieciatkowska4, J. Wade Harper2, and Samara L. Reck-Peterson1,3,5 1Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA, 92093. 2 Department of Cell Biology, Harvard Medical School, Boston, MA 02115. 3Section of Cell and Developmental Biology, Division of Biological Sciences, University of California San Diego, La Jolla, CA 92093. 4Department of Biochemistry and Molecular Genetics, University of Colorado Denver, Aurora, CO 80045. 5Howard Hughes Medical Institute †Present address: Stowers Institute for Medical Research, Kansas City, MO 64110 ‡Present address: Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138. *Correspondence to: Samara Reck-Peterson 9500 Gilman Drive, Leichtag 482 La Jolla CA, 92093 [email protected]; https://orcid.org/0000-0002-1553-465X 1 bioRxiv preprint doi: https://doi.org/10.1101/508887; this version posted December 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Product Datasheet KIF1C Antibody NB100-57510
Product Datasheet KIF1C Antibody NB100-57510 Unit Size: 0.1 ml Store at 4C. Do not freeze. Reviews: 1 Publications: 1 Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NB100-57510 Updated 3/16/2021 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NB100-57510 Page 1 of 3 v.20.1 Updated 3/16/2021 NB100-57510 KIF1C Antibody Product Information Unit Size 0.1 ml Concentration 0.2 mg/ml Storage Store at 4C. Do not freeze. Clonality Polyclonal Preservative 0.09% Sodium Azide Isotype IgG Purity Immunogen affinity purified Buffer TBS and 0.1% BSA Product Description Host Rabbit Gene ID 10749 Gene Symbol KIF1C Species Human, Mouse Immunogen The immunogen recognized by this antibody maps to a region between residue 1053 and the C-terminus (residue 1103) of human kinesin family member 1C (lethal toxin sensitivity 1) using the numbering given in entry NP_006603.2 (GeneID 10749). Product Application Details Applications Western Blot, Immunoprecipitation Recommended Dilutions Western Blot 1:2000-1:10000, Immunoprecipitation 2 - 5 ug/mg lysate Page 2 of 3 v.20.1 Updated 3/16/2021 Images Western Blot: KIF1C Antibody [NB100-57510] - KIF1C is a novel HOOK3 -interacting protein.sfGFP-3xFLAG and full length (FL) HOOK3, HOOK3 (1-552), and HOOK3 (553-718) all tagged with sfGFP and 3xFLAG were immunoprecipitated with alpha-FLAG antibodies from transiently transfected HEK-293T cells. Western blots with alpha-HC, alpha- FAM160A2, alpha-KIF1C, and alpha-FLAG antibodies were used to determine which proteins co-immunoprecipitated with each HOOK3 construct.DOI:http://dx.doi.org/10.7554/eLife.28257.015 Image collected and cropped by CiteAb from the following publication (https://elifesciences.org/articles/28257), licensed under a CC-BY licence. -

047605V1.Full.Pdf
bioRxiv preprint doi: https://doi.org/10.1101/047605; this version posted April 8, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Assembly and Activation of Dynein-Dynactin by the Cargo Adaptor Protein Hook3 Courtney M. Schroeder1,2 and Ronald D. Vale1,2 1The Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, California, USA 2Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, California, USA. Corresponding Author: Ronald D. Vale Dept. of Cellular and Molecular Pharmacology University of California, San Francisco Genentech Hall, MC 2200, Room N312A 600-16th Street San Francisco, CA 94158-2517 E-mail: [email protected] Phone: 415-476-6380 Fax: 415-514-4412 bioRxiv preprint doi: https://doi.org/10.1101/047605; this version posted April 8, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 2 Abstract Metazoan cytoplasmic dynein moves processively along microtubules with the aid of dynactin and an adaptor protein that joins dynein and dynactin into a stable ternary complex. Here, we have examined how Hook3, a cargo adaptor involved in Golgi and endosome transport, forms a motile dynein-dynactin complex. We show that the conserved Hook domain interacts directly with the dynein light intermediate chain 1 (LIC1). -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

Integrated Analysis of Germline and Tumor DNA Identifies New Candidate Genes Involved in Familial Colorectal Cancer
Supplementary Materials: Integrated Analysis of Germline and Tumor DNA Identifies New Candidate Genes Involved in Familial Colorectal Cancer Marcos Díaz-Gay, Sebastià Franch-Expósito, Coral Arnau-Collell, Solip Park, Fran Supek, Jenifer Muñoz, Laia Bonjoch, Anna Gratacós-Mulleras, Paula A. Sánchez-Rojas, Clara Esteban-Jurado, Teresa Ocaña, Miriam Cuatrecasas, Maria Vila-Casadesús, Juan José Lozano, Genis Parra, Steve Laurie, Sergi Beltran, EPICOLON Consortium, Antoni Castells, Luis Bujanda, Joaquín Cubiella, Francesc Balaguer and Sergi Castellví-Bel 100 80 10x ≥ age r 60 egions with cove r 40 ed r % of sha 20 0 I140 H458 H460 H461 H466 H468 H469 H470 FAM1 FAM3 FAM4 FAM19 FAM20 FAM22 FAM23 FAMN1 FAMN3 FAMN4 Families Figure S1. Histogram representing the percentage of genomic regions with a high-quality value of coverage (≥10×) with respect to all shared sequenced regions for each of the germline-tumor paired samples. Horizontal red line indicates sample filtering threshold (≥70% of shared regions with coverage above 10×). Figure S2. Pedigrees of the 18 families included in the study. Sample selected for germline and tumor whole-exome sequencing is indicated with an arrow. Filled symbols indicate affected for colorectal cancer (upper right quarter), adenoma/s (lower right quarter), gynecological cancer (ovary, uterine or breast cancer) (upper left quarter) and liver, stomach or pancreatic cancer (lower left quarter). Other cancer types are indicated in text with no symbol. IDs from samples undergoing germline whole-exome sequencing are also shown. AA/on-AA, advanced adenoma/non-advanced adenoma. Table S1. Description of germline copy number variants detected after calling with CoNIFER and ExomeDepth. -
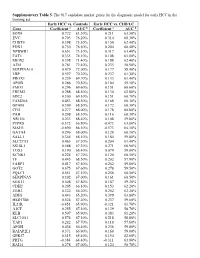
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

RNA Editing at Baseline and Following Endoplasmic Reticulum Stress
RNA Editing at Baseline and Following Endoplasmic Reticulum Stress By Allison Leigh Richards A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Human Genetics) in The University of Michigan 2015 Doctoral Committee: Professor Vivian G. Cheung, Chair Assistant Professor Santhi K. Ganesh Professor David Ginsburg Professor Daniel J. Klionsky Dedication To my father, mother, and Matt without whom I would never have made it ii Acknowledgements Thank you first and foremost to my dissertation mentor, Dr. Vivian Cheung. I have learned so much from you over the past several years including presentation skills such as never sighing and never saying “as you can see…” You have taught me how to think outside the box and how to create and explain my story to others. I would not be where I am today without your help and guidance. Thank you to the members of my dissertation committee (Drs. Santhi Ganesh, David Ginsburg and Daniel Klionsky) for all of your advice and support. I would also like to thank the entire Human Genetics Program, and especially JoAnn Sekiguchi and Karen Grahl, for welcoming me to the University of Michigan and making my transition so much easier. Thank you to Michael Boehnke and the Genome Science Training Program for supporting my work. A very special thank you to all of the members of the Cheung lab, past and present. Thank you to Xiaorong Wang for all of your help from the bench to advice on my career. Thank you to Zhengwei Zhu who has helped me immensely throughout my thesis even through my panic. -

Cargo Specific Regulation of Cytoplasmic Dynein by Effector Proteins
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2018 Cargo Specific Regulation Of Cytoplasmic Dynein By Effector Proteins Mara Olenick University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Biochemistry Commons, Biophysics Commons, and the Cell Biology Commons Recommended Citation Olenick, Mara, "Cargo Specific Regulation Of Cytoplasmic Dynein By Effector Proteins" (2018). Publicly Accessible Penn Dissertations. 3167. https://repository.upenn.edu/edissertations/3167 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/3167 For more information, please contact [email protected]. Cargo Specific Regulation Of Cytoplasmic Dynein By Effector Proteins Abstract Axonal transport is vital for the development and survival of neurons. The transport of cargo and organelles from the axon to the cell body is driven almost completely by the molecular motor, cytoplasmic dynein. Yet, it remains unclear how dynein is spatially and temporally regulated given the variety of cargo that must be properly localized to maintain cellular function. Previous work has suggested that adaptor proteins provide a mechanism for cargo-specific egulationr of motors. During my thesis work, I have investigated the role of mammalian Hook proteins, Hook1 and Hook3, as potential motor adaptors. Using optogenetic and single molecule assays, I found that Hook proteins interact with both dynein and dynactin, to effectively activate dynein motility, inducing longer run lengths and higher velocities than the previously characterized dynein activator, BICD2. In addition, I found that complex formation requires the N-terminal domain of Hook proteins, which resembles the calponin-homology domain of EB proteins yet cannot bind directly to microtubules. -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice. -

Structural Characterization of the RH1-LZI Tandem of JIP3/4
www.nature.com/scientificreports OPEN Structural characterization of the RH1-LZI tandem of JIP3/4 highlights RH1 domains as a cytoskeletal motor-binding motif Fernando Vilela1, Christophe Velours1, Mélanie Chenon1, Magali Aumont-Nicaise1, Valérie Campanacci1, Aurélien Thureau2, Olena Pylypenko3, Jessica Andreani 1, Paola Llinas1* & Julie Ménétrey 1* JIP3 and JIP4 (JNK-interacting proteins 3 and 4) are adaptors for cargo recruitment by dynein/dynactin and kinesin1 motors. Both are dimers that are stabilised by two sections of leucine zipper coiled coils. The N-terminal Leucine Zipper I (LZI) belongs to a section that binds dynein-DLIC and kinesin1-KHC, whilst the medial Leucine Zipper II (LZII) binds dynactin-p150glued and kinesin1-KLC. Structural data is available for the LZII, but the LZI section is still uncharacterized. Here we characterize the N-terminal part of JIP3/4 which consists of an RH1 (RILP homology 1) domain followed by the LZI coiled coil using bioinformatical, biophysical and structural approaches. The RH1-LZI tandem of JIP3 associates as a high afnity homodimer exhibiting elongated alpha-helical fold. 3D homology modelling of the RH1-LZI tandem reveals that the kinesin1-KHC binding site mainly overlaps with the RH1 domain. A sequence comparison search indicates that only one other protein family has RH1 domains similar to those of JIP3/4, the RILP (Rab-interacting lysosomal protein) family which consists of adaptor proteins linking Rab GTPases to cytoskeletal motors. RILPL2 is recruited through its RH1 domain by the myosin 5a motor. Here, we showed that the RH1 domain of JIP3 also interacts with myosin 5 A in vitro, highlighting JIP3/4 as possible myosin 5a adaptors. -

The Kinesin Superfamily Handbook Transporter, Creator, Destroyer
The Kinesin Superfamily Handbook Transporter, Creator, Destroyer Edited by Claire T. Friel First edition published 2020 ISBN: 978-1-138-58956-8 (hbk) ISBN: 978-0-429-49155-9 (ebk) 4 The Kinesin-3 Family Long-Distance Transporters Nida Siddiqui and Anne Straube CC BY-NC-ND 4.0 The Kinesin Superfamily Handbook The Kinesin-3 Family 4 Long-Distance Transporters Nida Siddiqui and Anne Straube CONTENTS 4.1 Example Family Members .............................................................................. 41 4.2 Structural Information .................................................................................... 41 4.3 Functional Properties ...................................................................................... 43 4.3.1 Autoinhibition of Kinesin-3 Motors and Their Activation .................45 4.4 Physiological Roles .........................................................................................46 4.4.1 Preference for Subsets of Microtubule Tracks .................................... 47 4.5 Involvement in Disease ...................................................................................48 Acknowledgements ..................................................................................................49 References ................................................................................................................49 The Kinesin-3s are a family of cargo transporters. They typically display highly processive plus-end-directed motion, either as dimers or in teams, formed via interaction with