Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

Mutant IDH, (R)-2-Hydroxyglutarate, and Cancer
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer Julie-Aurore Losman1 and William G. Kaelin Jr.1,2,3 1Department of Medical Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02215, USA; 2Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA Mutations in metabolic enzymes, including isocitrate whether altered cellular metabolism is a cause of cancer dehydrogenase 1 (IDH1) and IDH2, in cancer strongly or merely an adaptive response of cancer cells in the face implicate altered metabolism in tumorigenesis. IDH1 of accelerated cell proliferation is still a topic of some and IDH2 catalyze the interconversion of isocitrate and debate. 2-oxoglutarate (2OG). 2OG is a TCA cycle intermediate The recent identification of cancer-associated muta- and an essential cofactor for many enzymes, including tions in three metabolic enzymes suggests that altered JmjC domain-containing histone demethylases, TET cellular metabolism can indeed be a cause of some 5-methylcytosine hydroxylases, and EglN prolyl-4-hydrox- cancers (Pollard et al. 2003; King et al. 2006; Raimundo ylases. Cancer-associated IDH mutations alter the enzymes et al. 2011). Two of these enzymes, fumarate hydratase such that they reduce 2OG to the structurally similar (FH) and succinate dehydrogenase (SDH), are bone fide metabolite (R)-2-hydroxyglutarate [(R)-2HG]. Here we tumor suppressors, and loss-of-function mutations in FH review what is known about the molecular mechanisms and SDH have been identified in various cancers, in- of transformation by mutant IDH and discuss their im- cluding renal cell carcinomas and paragangliomas. -

Supplemental Material
Supplemental Table B ARGs in alphabetical order Symbol Title 3 months 6 months 9 months 12 months 23 months ANOVA Direction Category 38597 septin 2 1557 ± 44 1555 ± 44 1579 ± 56 1655 ± 26 1691 ± 31 0.05219 up Intermediate 0610031j06rik kidney predominant protein NCU-G1 491 ± 6 504 ± 14 503 ± 11 527 ± 13 534 ± 12 0.04747 up Early Adult 1G5 vesicle-associated calmodulin-binding protein 662 ± 23 675 ± 17 629 ± 16 617 ± 20 583 ± 26 0.03129 down Intermediate A2m alpha-2-macroglobulin 262 ± 7 272 ± 8 244 ± 6 290 ± 7 353 ± 16 0.00000 up Midlife Aadat aminoadipate aminotransferase (synonym Kat2) 180 ± 5 201 ± 12 223 ± 7 244 ± 14 275 ± 7 0.00000 up Early Adult Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 958 ± 28 1052 ± 58 1086 ± 36 1071 ± 44 1141 ± 41 0.05371 up Early Adult Abcb1a ATP-binding cassette, sub-family B (MDR/TAP), member 1A 136 ± 8 147 ± 6 147 ± 13 155 ± 9 185 ± 13 0.01272 up Midlife Acadl acetyl-Coenzyme A dehydrogenase, long-chain 423 ± 7 456 ± 11 478 ± 14 486 ± 13 512 ± 11 0.00003 up Early Adult Acadvl acyl-Coenzyme A dehydrogenase, very long chain 426 ± 14 414 ± 10 404 ± 13 411 ± 15 461 ± 10 0.01017 up Late Accn1 amiloride-sensitive cation channel 1, neuronal (degenerin) 242 ± 10 250 ± 9 237 ± 11 247 ± 14 212 ± 8 0.04972 down Late Actb actin, beta 12965 ± 310 13382 ± 170 13145 ± 273 13739 ± 303 14187 ± 269 0.01195 up Midlife Acvrinp1 activin receptor interacting protein 1 304 ± 18 285 ± 21 274 ± 13 297 ± 21 341 ± 14 0.03610 up Late Adk adenosine kinase 1828 ± 43 1920 ± 38 1922 ± 22 2048 ± 30 1949 ± 44 0.00797 up Early -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

The Role of EZH2 in the Induction and Maintenance of Acute Myeloid Leukaemia
The Role of EZH2 in the Induction and Maintenance of Acute Myeloid Leukaemia Faisal Basheer Downing College University of Cambridge This dissertation is submitted for the degree of Doctor of Philosophy September 2017 Declaration The work presented in this dissertation was carried out under the supervision of Professor Brian Huntly from November 2013 to November 2016 in the Department of Haematology, University of Cambridge. This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except as declared in the Preface and specified in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. It does not exceed the prescribed word limit set by the Clinical Medicine and Clinical Veterinary Medicine Degree Committee. Faisal Basheer September 2017 i Acknowledgements First and foremost, I would like to thank my supervisor, Professor Brian Huntly, for providing me with the opportunity to work towards this thesis - for his help, encouragement and support over the last four years of my PhD. His dedicated high-level supervision, meticulous attention to detail and excellent direction throughout this time has undoubtedly helped me to maximise my potential in the lab throughout a highly interesting and rewarding project. I would also like to thank all of my colleagues and friends in the Huntly laboratory over the past four years - working together with all of you as a team has been an unforgettable and rewarding experience. -
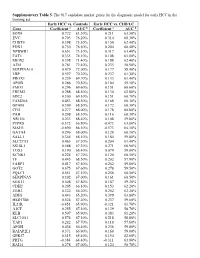
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

High Density Genome Wide Genotyping-By-Sequencing and Association
www.nature.com/scientificreports OPEN High density genome wide genotyping-by-sequencing and association identifies common and Received: 24 March 2016 Accepted: 14 July 2016 low frequency SNPs, and novel Published: 10 August 2016 candidate genes influencing cow milk traits Eveline M. Ibeagha-Awemu1, Sunday O. Peters2, Kingsley A. Akwanji3, Ikhide G. Imumorin4 & Xin Zhao3 High-throughput sequencing technologies have increased the ability to detect sequence variations for complex trait improvement. A high throughput genome wide genotyping-by-sequencing (GBS) method was used to generate 515,787 single nucleotide polymorphisms (SNPs), from which 76,355 SNPs with call rates >85% and minor allele frequency ≥1.5% were used in genome wide association study (GWAS) of 44 milk traits in 1,246 Canadian Holstein cows. GWAS was accomplished with a mixed linear model procedure implementing the additive and dominant models. A strong signal within the centromeric region of bovine chromosome 14 was associated with test day fat percentage. Several SNPs were associated with eicosapentaenoic acid, docosapentaenoic acid, arachidonic acid, CLA:9c11t and gamma linolenic acid. Most of the significant SNPs for 44 traits studied are novel and located in intergenic regions or introns of genes. Novel potential candidate genes for milk traits or mammary gland functions include ERCC6, TONSL, NPAS2, ACER3, ITGB4, GGT6, ACOX3, MECR, ADAM12, ACHE, LRRC14, FUK, NPRL3, EVL, SLCO3A1, PSMA4, FTO, ADCK5, PP1R16A and TEP1. Our study further demonstrates the utility of the GBS approach for identifying population-specific SNPs for use in improvement of complex dairy traits. Milk is an important source of nutrients in human nutrition all over the world. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Regulators of G Protein Signaling in Analgesia and Addiction
Molecular Pharmacology Fast Forward. Published on May 30, 2020 as DOI: 10.1124/mol.119.119206MOL Manuscript # 119206 This article has not been copyedited and formatted. The final version may differ from this version. Regulators of G protein signaling in analgesia and addiction Farhana Sakloth1, Claire Polizu1, Feodora Bertherat1, and Venetia Zachariou1,2 1Nash Family Department of Neuroscience, and Friedman Brain Institute, Icahn School of Downloaded from Medicine at Mount Sinai, 1425 Madison Ave, New York, NY, 10029. 2 Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, 1425 molpharm.aspetjournals.org Madison Ave, New York, NY, 10029. at ASPET Journals on October 6, 2021 Abbreviated title: RGS proteins in analgesia and addiction Corresponding author: Venetia Zachariou, Icahn School of Medicine at Mount Sinai, 1425 Madison Ave, New York, NY, 10029. Phone: 212-6598612, Email: [email protected] 1 Molecular Pharmacology Fast Forward. Published on May 30, 2020 as DOI: 10.1124/mol.119.119206MOL Manuscript # 119206 This article has not been copyedited and formatted. The final version may differ from this version. Pages: 39 Tables: 1 Figures: 3 References: 131 Abstract: 219 Introduction: 754 Discussion: 257 Downloaded from molpharm.aspetjournals.org Keywords: Signal transduction, GPCR, reward, analgesic tolerance, physical dependence, RGS, cocaine, amphetamine at ASPET Journals on October 6, 2021 2 Molecular Pharmacology Fast Forward. Published on May 30, 2020 as DOI: 10.1124/mol.119.119206MOL Manuscript # 119206 This article has not been copyedited and formatted. The final version may differ from this version. ABSTRACT Regulators of G protein signaling (RGS) are multifunctional proteins expressed in peripheral and neuronal cells, playing critical roles in development, physiological processes, and pharmacological responses. -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice.