Supplemental Material
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

Unnatural Verticilide Enantiomer Inhibits Type 2 Ryanodine Receptor-Mediated Calcium Leak and Is Antiarrhythmic
Unnatural verticilide enantiomer inhibits type 2 ryanodine receptor-mediated calcium leak and is antiarrhythmic Suzanne M. Batistea,1, Daniel J. Blackwellb,1, Kyungsoo Kimb,1, Dmytro O. Kryshtalb, Nieves Gomez-Hurtadob, Robyn T. Rebbeckc, Razvan L. Corneac, Jeffrey N. Johnstona,2, and Bjorn C. Knollmannb,2 aDepartment of Chemistry, Vanderbilt University, Nashville, TN 37235; bDepartment of Medicine, Vanderbilt University Medical Center, Nashville, TN 37232; and cDepartment of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, MN 55455 Edited by Dale L. Boger, The Scripps Research Institute, La Jolla, CA, and approved January 15, 2019 (received for review September 27, 2018) Ca2+ leak via ryanodine receptor type 2 (RyR2) can cause poten- heart diseases associated with both atrial and ventricular arrhyth- tially fatal arrhythmias in a variety of heart diseases and has also mia (9). Mutations in RyR2 and its binding partners, which increase + been implicated in neurodegenerative and seizure disorders, mak- SR Ca2 leak, cause primary atrial and ventricular arrhythmia ing RyR2 an attractive therapeutic target for drug development. syndromes such as catecholaminergic polymorphic ventricular Here we synthesized and investigated the fungal natural product tachycardia (CPVT), providing strong evidence for the mechanistic and known insect RyR antagonist (−)-verticilide and several conge- contribution of RyR2 to arrhythmia risk in humans (10). Further ners to determine their activity against mammalian RyR2. Although support comes from gene-targeted mouse models of CPVT, where + the cyclooligomeric depsipeptide natural product (−)-verticilide had catecholamine-induced spontaneous Ca2 release from the SR no effect, its nonnatural enantiomer [ent-(+)-verticilide] signifi- via RyR2 generates potentially fatal cardiac arrhythmias (11, 12). -

PFN2, a Novel Marker of Unfavorable Prognosis, Is a Potential Therapeutic
Cui et al. J Transl Med (2016) 14:137 DOI 10.1186/s12967-016-0884-y Journal of Translational Medicine RESEARCH Open Access PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma Xiao‑bin Cui1,2†, Shu‑mao Zhang1†, Yue‑xun Xu3, Hong‑wei Dang1, Chun‑xia Liu1, Liang‑hai Wang1, Lan Yang1, Jian‑ming Hu1, Wei‑hua Liang1, Jin‑fang Jiang1, Na Li4, Yong Li5*, Yun‑zhao Chen1* and Feng Li1,2* Abstract Background: Esophageal squamous cell carcinoma (ESCC) is one of the most aggressively malignant tumors with dismal prognosis. Profilin 2 (PFN2) is an actin-binding protein that regulates the dynamics of actin polymerization and plays a key role in cell motility. Recently, PFN2 have emerged as significant regulators of cancer processes. However, the clinical significance and biological function of PFN2 in ESCC remain unclear. Methods: PFN2 protein expression was validated by immunohistochemistry (IHC) on tissue microarray from Chinese Han and Kazakh populations with ESCC. The associations among PFN2 expression, clinicopathological features, and prognosis of ESCC were analyzed. The effects on cell proliferation, invasion and migration were examined using MTT and Transwell assays. Markers of epithelial–mesenchymal transition (EMT) were detected by Western blot analysis. Results: Compared with normal esophageal epithelium (NEE), PFN2 protein expression was markedly increased in low-grade intraepithelial neoplasia (LGIN), high-grade intraepithelial neoplasia (HGIN), and ESCC, increased gradually from LGIN to ESCC, and finally reached high grade in HGIN in the Han population. Similarly, PFN2 protein was more overexpressed in ESCC than in NEE in the Kazakh population. -

Decreased Expression of Profilin 2 in Oral Squamous Cell Carcinoma and Its Clinicopathological Implications
ONCOLOGY REPORTS 26: 813-823, 2011 Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications C.Y. MA1,2, C.P. ZHANG1,2, L.P. ZHONG1,2, H.Y. PAN1,2, W.T. CHEN1,2, L.Z. WANG3, O.W. ANDREW4, T. JI1 and W. HAN1,2 1Department of Oral and Maxillofacial Surgery, Ninth People's Hospital, College of Stomatology; 2Shanghai Key Laboratory of Stomatology and Shanghai Research Institute of Stomatology; 3Department of Oral Pathology, Ninth People's Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, P.R. China; 4Department of Oral and Maxillofacial Surgery, Faculty of Dentistry, National University of Singapore, Singapore 119074, Singapore Received February 8, 2011; Accepted April 11, 2011 DOI: 10.3892/or.2011.1365 Abstract. Profilins are small proteins essential for many clinical and pathological significance. In conclusion, PFN2 normal cellular dynamics and constitute one of the crucial can be utilized as both a potential suppressor marker and a components of actin-based cellular motility. Several recent prognostic protein for OSCC. The function of PFN2 may be to studies have implicated a role for the profilin (PFN) family in regulate the N-WASP/Arp2/3 signaling pathway. cancer pathogenesis and progression. However, their expression and promising functions are largely unknown in oral squamous Introduction cell carcinoma (OSCC). In this study, we analyzed the correlation between PFN1 and PFN2 expression in vitro and Oral squamous cell carcinoma (OSCC) is a significant public in vivo. The protein expression levels were roughly compared health problem with >300,000 new cases being diagnosed between cell lines (HIOEC, HB96) with the employment of annually worldwide (1). -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

Aquaporin Channels in the Heart—Physiology and Pathophysiology
International Journal of Molecular Sciences Review Aquaporin Channels in the Heart—Physiology and Pathophysiology Arie O. Verkerk 1,2,* , Elisabeth M. Lodder 2 and Ronald Wilders 1 1 Department of Medical Biology, Amsterdam University Medical Centers, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; [email protected] 2 Department of Experimental Cardiology, Amsterdam University Medical Centers, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; [email protected] * Correspondence: [email protected]; Tel.: +31-20-5664670 Received: 29 March 2019; Accepted: 23 April 2019; Published: 25 April 2019 Abstract: Mammalian aquaporins (AQPs) are transmembrane channels expressed in a large variety of cells and tissues throughout the body. They are known as water channels, but they also facilitate the transport of small solutes, gasses, and monovalent cations. To date, 13 different AQPs, encoded by the genes AQP0–AQP12, have been identified in mammals, which regulate various important biological functions in kidney, brain, lung, digestive system, eye, and skin. Consequently, dysfunction of AQPs is involved in a wide variety of disorders. AQPs are also present in the heart, even with a specific distribution pattern in cardiomyocytes, but whether their presence is essential for proper (electro)physiological cardiac function has not intensively been studied. This review summarizes recent findings and highlights the involvement of AQPs in normal and pathological cardiac function. We conclude that AQPs are at least implicated in proper cardiac water homeostasis and energy balance as well as heart failure and arsenic cardiotoxicity. However, this review also demonstrates that many effects of cardiac AQPs, especially on excitation-contraction coupling processes, are virtually unexplored. -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
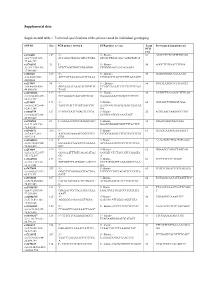
Supplemental Data Supplemental Table 1. Technical Specifications of the Primers Used for Individual Genotyping
Supplemental data Supplemental table 1. Technical specifications of the primers used for individual genotyping SNP ID Size PCR primer forward PCR primer reverse Temp Pyrosequencing primer(s) PCR (°C) rs776108 127 5’- 5’- Biotin- 61 ATACCTCTCATTTTGCAG chr3:77,825,927- ACCAGGCTAGGCATGCTATA GTCACTTAACAGCAGTGTGTCA 77,826,729 rs3746192 92 5’- 5’- Biotin- 56 AGGCTTGTAACCTGGA chr19:17,946,368- ATTCTAGGTGGCATGAGGG CTGGGGAGCAACAGAAGA 17,946,488 rs988147 169 5' - 5' - Biotine- 53 GCAGGGGGTAGAAATG chr6:45282108- AGCCATTAAAGAATTTCAAA TTGGATTTTATTCTTGTAATAGG 45282608 rs227849 94 5' - 5' - Biotine- 56 GGTTTAAGGTCTTTGCAT chr6:44,806,436- AGGAAAATAAACATGTGGTT TCTACCAATATTTTCTTTCGTAG 44,806,936 TAAG T rs10733833 127 5' - 5' - Biotine- 58 CATGTTTAAAACCTTTCAG chr10:68,418,227- GCCAAAACCAACAGTTCAT GAAAAAAATTGCACCTGTCTC 68,418,727 rs322609 197 5’- 5’-Biotin- 60 GGTAGCTGTGGGTGGA chr16:62,432,604- TAGTTGATTTTGCCAACCTG AAATGGGTGACAGAAGTAATAA 62,433,104 GA rs1884779 127 5’-TGGCTATTGGAGTTCTCA 5’-Biotin- 55 AGTGAATTAAGGGCTTGT chr20:45,857,969- CCATCCATCCCAAATAGT 45,858,469 rs4703908 83 5’- GAAAATGCCCAAGGTGAC 5’-Biotin- 52 GTGACAGTGGGCAAA chr5:71,802,353- GAATGTGGGTGTGTTTTACTCT 71,802,853 rs6946871 290 5’- 5’-Biotin- 61 GGAGGAAAGGAAAAGTT chr7:4,037,310- ATCAGATAAAATCGGCTTCT TCGGGAAGGTTTTTGTACTTTTG 4,037,810 GTG rs11865033 123 5’- 5’-Biotin- 61 AAAGTCTCTTCCTATGAGC chr16:78,082,945- AATAAACCAAGCCCTGAAAA ACTAAAATCCCCCTTTCCTCCA 78,083,445 GTC rs247004 170 5’- 5’-Biotin- 62 GGAAGCCAGACTAGCAG chr5:131,372,007- GGGGAATTTGTCAGAGATAG GGGATCCTCTACCATCCAAATA 131,372,507 GG -

Neuroprotective Effects of Geniposide from Alzheimer's Disease Pathology
Neuroprotective effects of geniposide from Alzheimer’s disease pathology WeiZhen Liu1, Guanglai Li2, Christian Hölscher2,3, Lin Li1 1. Key Laboratory of Cellular Physiology, Shanxi Medical University, Taiyuan, PR China 2. Second hospital, Shanxi medical University, Taiyuan, PR China 3. Neuroscience research group, Faculty of Health and Medicine, Lancaster University, Lancaster LA1 4YQ, UK running title: Neuroprotective effects of geniposide corresponding author: Prof. Lin Li Key Laboratory of Cellular Physiology, Shanxi Medical University, Taiyuan, PR China Email: [email protected] Neuroprotective effects of geniposide Abstract A growing body of evidence have linked two of the most common aged-related diseases, type 2 diabetes mellitus (T2DM) and Alzheimer disease (AD). It has led to the notion that drugs developed for the treatment of T2DM may be beneficial in modifying the pathophysiology of AD. As a receptor agonist of glucagon- like peptide (GLP-1R) which is a newer drug class to treat T2DM, Geniposide shows clear effects in inhibiting pathological processes underlying AD, such as and promoting neurite outgrowth. In the present article, we review possible molecular mechanisms of geniposide to protect the brain from pathologic damages underlying AD: reducing amyloid plaques, inhibiting tau phosphorylation, preventing memory impairment and loss of synapses, reducing oxidative stress and the chronic inflammatory response, and promoting neurite outgrowth via the GLP-1R signaling pathway. In summary, the Chinese herb geniposide shows great promise as a novel treatment for AD. Key words: Alzheimer’s disease, geniposide, amyloid-β, neurofibrillary tangles, oxidative stress, inflammatation, type 2 diabetes mellitus, glucagon like peptide receptor, neuroprotection, tau protein Neuroprotective effects of geniposide 1. -

Oxido-Reductive Regulation of Vascular Remodeling by Receptor Tyrosine Kinase ROS1
Oxido-reductive regulation of vascular remodeling by receptor tyrosine kinase ROS1 Ziad A. Ali, … , Thomas Quertermous, Euan A. Ashley J Clin Invest. 2014;124(12):5159-5174. https://doi.org/10.1172/JCI77484. Research Article Angioplasty and stenting is the primary treatment for flow-limiting atherosclerosis; however, this strategy is limited by pathological vascular remodeling. Using a systems approach, we identified a role for the network hub gene glutathione peroxidase-1 (GPX1) in pathological remodeling following human blood vessel stenting. Constitutive deletion ofG px1 in atherosclerotic mice recapitulated this phenotype of increased vascular smooth muscle cell (VSMC) proliferation and plaque formation. In an independent patient cohort, gene variant pair analysis identified an interaction of GPX1 with the orphan protooncogene receptor tyrosine kinase ROS1. A meta-analysis of the only genome-wide association studies of human neointima-induced in-stent stenosis confirmed the association of the ROS1 variant with pathological remodeling. Decreased GPX1 expression in atherosclerotic mice led to reductive stress via a time-dependent increase in glutathione, corresponding to phosphorylation of the ROS1 kinase activation site Y2274. Loss of GPX1 function was associated with both oxidative and reductive stress, the latter driving ROS1 activity via s-glutathiolation of critical residues of the ROS1 tyrosine phosphatase SHP-2. ROS1 inhibition with crizotinib and deglutathiolation of SHP-2 abolished GPX1-mediated increases in VSMC proliferation while leaving endothelialization intact. Our results indicate that GPX1-dependent alterations in oxido-reductive stress promote ROS1 activation and mediate vascular remodeling. Find the latest version: https://jci.me/77484/pdf The Journal of Clinical Investigation RESEARCH ARTICLE Oxido-reductive regulation of vascular remodeling by receptor tyrosine kinase ROS1 Ziad A. -

Tropomyosin Receptor Antagonism in Cylindromatosis (TRAC), an Early Phase Trial of a Topical Tropomyosin Kinase Inhibitor As
Cranston et al. Trials (2017) 18:111 DOI 10.1186/s13063-017-1812-z STUDY PROTOCOL Open Access Tropomyosin Receptor Antagonism in Cylindromatosis (TRAC), an early phase trial of a topical tropomyosin kinase inhibitor as a treatment for inherited CYLD defective skin tumours: study protocol for a randomised controlled trial Amy Cranston1* , Deborah D. Stocken1,2, Elaine Stamp2, David Roblin3, Julia Hamlin4, James Langtry5, Ruth Plummer6, Alan Ashworth7, John Burn8 and Neil Rajan5,8 Abstract Background: Patients with germline mutations in a tumour suppressor gene called CYLD develop multiple, disfiguring, hair follicle tumours on the head and neck. The prognosis is poor, with up to one in four mutation carriers requiring complete surgical removal of the scalp. There are no effective medical alternatives to treat this condition. Whole genome molecular profiling experiments led to the discovery of an attractive molecular target in these skin tumour cells, named tropomyosin receptor kinase (TRK), upon which these cells demonstrate an oncogenic dependency in preclinical studies. Recently, the development of an ointment containing a TRK inhibitor (pegcantratinib — previously CT327 — from Creabilis SA) allowed for the assessment of TRK inhibition in tumours from patients with inherited CYLD mutations. Methods/design: Tropomysin Receptor Antagonism in Cylindromatosis (TRAC) is a two-part, exploratory, early phase, single-centre trial. Cohort 1 is a phase 1b open-labelled trial, and cohort 2 is a phase 2a randomised double-blinded exploratory placebo-controlled trial. Cohort 1 will determine the safety and acceptability of applying pegcantratinib for 4 weeks to a single tumour on a CYLD mutation carrier that is scheduled for a routine lesion excision (n = 8 patients).