Decreased Expression of Profilin 2 in Oral Squamous Cell Carcinoma and Its Clinicopathological Implications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

PFN2, a Novel Marker of Unfavorable Prognosis, Is a Potential Therapeutic
Cui et al. J Transl Med (2016) 14:137 DOI 10.1186/s12967-016-0884-y Journal of Translational Medicine RESEARCH Open Access PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma Xiao‑bin Cui1,2†, Shu‑mao Zhang1†, Yue‑xun Xu3, Hong‑wei Dang1, Chun‑xia Liu1, Liang‑hai Wang1, Lan Yang1, Jian‑ming Hu1, Wei‑hua Liang1, Jin‑fang Jiang1, Na Li4, Yong Li5*, Yun‑zhao Chen1* and Feng Li1,2* Abstract Background: Esophageal squamous cell carcinoma (ESCC) is one of the most aggressively malignant tumors with dismal prognosis. Profilin 2 (PFN2) is an actin-binding protein that regulates the dynamics of actin polymerization and plays a key role in cell motility. Recently, PFN2 have emerged as significant regulators of cancer processes. However, the clinical significance and biological function of PFN2 in ESCC remain unclear. Methods: PFN2 protein expression was validated by immunohistochemistry (IHC) on tissue microarray from Chinese Han and Kazakh populations with ESCC. The associations among PFN2 expression, clinicopathological features, and prognosis of ESCC were analyzed. The effects on cell proliferation, invasion and migration were examined using MTT and Transwell assays. Markers of epithelial–mesenchymal transition (EMT) were detected by Western blot analysis. Results: Compared with normal esophageal epithelium (NEE), PFN2 protein expression was markedly increased in low-grade intraepithelial neoplasia (LGIN), high-grade intraepithelial neoplasia (HGIN), and ESCC, increased gradually from LGIN to ESCC, and finally reached high grade in HGIN in the Han population. Similarly, PFN2 protein was more overexpressed in ESCC than in NEE in the Kazakh population. -

In Situlocalization of Cytoskeletal Elements in the Human Trabecular
Investigative Ophthalmology & Visual Science. Vol. 31. No. 9. September 1990 Cops right £• Association lor Research in Vision and Ophthalmology In Situ Localization of Cytoskeletal Elements in the Human Trabecular Meshwork and Cornea Robert N. Weinreb* and Mark I. Ryderf The authors compared cytoskeletal elements of the in situ human trabccular-mcshwork cell with in situ human corneal cells using indirect immunofluorcsccncc staining for tubulin and intermediate filaments (vimentin, cytokeratin, and desmin) and NBD-phallacidin staining for f-actin using both fixed frozen and unfixed frozen sections from postmortem eyes. Both f-actin and tubulin were found throughout the cell body of trabecular-meshwork cells, keratocytes, corneal endothelium, and corneal epithelium. The f-actin staining pattern was concentrated at the cell periphery of these four cell types. Vimentin stain was intensely localized in focal areas of the trabecular-meshwork cell, keratocytes, and throughout the corneal cndothelium. A general anticytokeratin antibody was intensely localized in corneal epithelium and endothelium. However, PKK-1 anticytokeratin antibody was seen only in superficial layers of corneal epithelium and not in corneal endothelium. The 4.62 anticytokeratin antibody was not observed in either corneal epithelium or endothelium. None of these three cytokera- tin antibodies were seen in trabccular-mcshwork cells or keratocytes. Desmin stain was not noted in any of these cell types. In general, cytoskeletal staining of unfixed frozen sections showed a similar staining pattern for f-actin and tubulin but a more uniform and intense staining pattern for vimentin and cytokcratin compared with fixed frozen material. The authors conclude that these cytoskclctal stains can differentiate human Irabeciilar-meshwork cells from cells of the cornea in situ. -

Genome-Wide Analyses Identify KIF5A As a Novel ALS Gene
This is a repository copy of Genome-wide Analyses Identify KIF5A as a Novel ALS Gene.. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/129590/ Version: Accepted Version Article: Nicolas, A, Kenna, KP, Renton, AE et al. (210 more authors) (2018) Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron, 97 (6). 1268-1283.e6. https://doi.org/10.1016/j.neuron.2018.02.027 Reuse This article is distributed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs (CC BY-NC-ND) licence. This licence only allows you to download this work and share it with others as long as you credit the authors, but you can’t change the article in any way or use it commercially. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Genome-wide Analyses Identify KIF5A as a Novel ALS Gene Aude Nicolas1,2, Kevin P. Kenna2,3, Alan E. Renton2,4,5, Nicola Ticozzi2,6,7, Faraz Faghri2,8,9, Ruth Chia1,2, Janice A. Dominov10, Brendan J. Kenna3, Mike A. Nalls8,11, Pamela Keagle3, Alberto M. Rivera1, Wouter van Rheenen12, Natalie A. Murphy1, Joke J.F.A. van Vugt13, Joshua T. Geiger14, Rick A. Van der Spek13, Hannah A. Pliner1, Shankaracharya3, Bradley N. -

Supplemental Material
Supplemental Table B ARGs in alphabetical order Symbol Title 3 months 6 months 9 months 12 months 23 months ANOVA Direction Category 38597 septin 2 1557 ± 44 1555 ± 44 1579 ± 56 1655 ± 26 1691 ± 31 0.05219 up Intermediate 0610031j06rik kidney predominant protein NCU-G1 491 ± 6 504 ± 14 503 ± 11 527 ± 13 534 ± 12 0.04747 up Early Adult 1G5 vesicle-associated calmodulin-binding protein 662 ± 23 675 ± 17 629 ± 16 617 ± 20 583 ± 26 0.03129 down Intermediate A2m alpha-2-macroglobulin 262 ± 7 272 ± 8 244 ± 6 290 ± 7 353 ± 16 0.00000 up Midlife Aadat aminoadipate aminotransferase (synonym Kat2) 180 ± 5 201 ± 12 223 ± 7 244 ± 14 275 ± 7 0.00000 up Early Adult Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 958 ± 28 1052 ± 58 1086 ± 36 1071 ± 44 1141 ± 41 0.05371 up Early Adult Abcb1a ATP-binding cassette, sub-family B (MDR/TAP), member 1A 136 ± 8 147 ± 6 147 ± 13 155 ± 9 185 ± 13 0.01272 up Midlife Acadl acetyl-Coenzyme A dehydrogenase, long-chain 423 ± 7 456 ± 11 478 ± 14 486 ± 13 512 ± 11 0.00003 up Early Adult Acadvl acyl-Coenzyme A dehydrogenase, very long chain 426 ± 14 414 ± 10 404 ± 13 411 ± 15 461 ± 10 0.01017 up Late Accn1 amiloride-sensitive cation channel 1, neuronal (degenerin) 242 ± 10 250 ± 9 237 ± 11 247 ± 14 212 ± 8 0.04972 down Late Actb actin, beta 12965 ± 310 13382 ± 170 13145 ± 273 13739 ± 303 14187 ± 269 0.01195 up Midlife Acvrinp1 activin receptor interacting protein 1 304 ± 18 285 ± 21 274 ± 13 297 ± 21 341 ± 14 0.03610 up Late Adk adenosine kinase 1828 ± 43 1920 ± 38 1922 ± 22 2048 ± 30 1949 ± 44 0.00797 up Early -
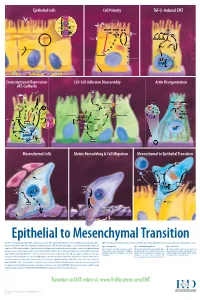
Epithelial to Mesenchymal Transition
Epithelial Cells Cell Polarity TGF-b-Induced EMT MUC-1 O-glycosylation Epithelial Cells ZO-1 Occludin Apical Membrane Tight F-Actin Microvilli Junction Claudin F-Actin p120 β-Catenin Adherens F-Actin Ezrin TGF-β dimer Junction E-Cadherin α-Catenin Plakophilin Crumbs Complex PAR Complex Desmocollin Desmoplakin Desmosome PtdIns(4,5)P2 TGF-β RII TGF-β RI CRB Cdc42Par6 Desmoglein Cytokeratin Pals1 PatJ Tight Junction Plakoglobin aPKC Par3 Domain Smad7 Extracellular PTEN JNK ERK1/2 p38 SARA Smurf1 Cortical Actin Cytoskeleton Space Par3 ZO-1 Adherens Junction PI 3-K Domain Smad-independent Signaling (–) Smad7 Translocation Smad2/3 PtdIns(3,4,5)P3 Smad4 Smad4 NEDD4 Cytokeratin Intermediate Filaments Smad2 Smad4 Smad3 LLGL Proteasome SCRIB DLG Scribble Complex Fibronectin Twist Smad2/3 Vitronectin ZEB 1/2 Microtubule Network Smad4 N-Cadherin Snail Basolateral Membrane CoA, Collagen I Slug CoR MMPs DNA-binding (+) Claudin Desmoplakin Transcription Factor Occludin Cytokeratins E-Cadherin Plakoglobin Integrins β α Nidogen-1/Entactin Perlecan Laminin Collagen IV Transcriptional Repression Cell-Cell Adhesion Disassembly Actin Reorganization of E-Cadherin TGF-β dimer EGF TGF-β RII TGF-β RI IGF FGF Receptor TNF-α Tyrosine Kinase Par6 TNF RI Apical Focal Adhesion Constriction Actin Depolymerization F-Actin Smurf1 Occludin Wnt Frizzled Myosin II Ras RhoA α-Actinin Myosin II ROCK AxinCK1 Dishevelled GSK-3 PI 3-K Src Zyxin MLC Phosphatase APC Proteasome FAK Vinculin RhoA ILK Talin (Inactive) Hakai Talin FAK F-Actin E-Cadherin LIMK Akt Paxillin FAK Stress -

Serum Albumin OS=Homo Sapiens
Protein Name Cluster of Glial fibrillary acidic protein OS=Homo sapiens GN=GFAP PE=1 SV=1 (P14136) Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 Cluster of Isoform 3 of Plectin OS=Homo sapiens GN=PLEC (Q15149-3) Cluster of Hemoglobin subunit beta OS=Homo sapiens GN=HBB PE=1 SV=2 (P68871) Vimentin OS=Homo sapiens GN=VIM PE=1 SV=4 Cluster of Tubulin beta-3 chain OS=Homo sapiens GN=TUBB3 PE=1 SV=2 (Q13509) Cluster of Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 (P60709) Cluster of Tubulin alpha-1B chain OS=Homo sapiens GN=TUBA1B PE=1 SV=1 (P68363) Cluster of Isoform 2 of Spectrin alpha chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTAN1 (Q13813-2) Hemoglobin subunit alpha OS=Homo sapiens GN=HBA1 PE=1 SV=2 Cluster of Spectrin beta chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTBN1 PE=1 SV=2 (Q01082) Cluster of Pyruvate kinase isozymes M1/M2 OS=Homo sapiens GN=PKM PE=1 SV=4 (P14618) Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 Clathrin heavy chain 1 OS=Homo sapiens GN=CLTC PE=1 SV=5 Filamin-A OS=Homo sapiens GN=FLNA PE=1 SV=4 Cytoplasmic dynein 1 heavy chain 1 OS=Homo sapiens GN=DYNC1H1 PE=1 SV=5 Cluster of ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide OS=Homo sapiens GN=ATP1A2 PE=3 SV=1 (B1AKY9) Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 Dihydropyrimidinase-related protein 2 OS=Homo sapiens GN=DPYSL2 PE=1 SV=1 Cluster of Alpha-actinin-1 OS=Homo sapiens GN=ACTN1 PE=1 SV=2 (P12814) 60 kDa heat shock protein, mitochondrial OS=Homo -

Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations
Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations by Jelena Perovanovic M.S. in Molecular Biology and Physiology, September 2009, University of Belgrade M.Phil. in Molecular Medicine, August 2013, The George Washington University A Dissertation submitted to The Faculty of The Columbian College of Arts and Sciences of The George Washington University in partial fulfillment of the requirements for the degree of Doctor of Philosophy August 31, 2015 Dissertation directed by Eric P. Hoffman Professor of Integrative Systems Biology The Columbian College of Arts and Sciences of The George Washington University certifies that Jelena Perovanovic has passed the Final Examination for the degree of Doctor of Philosophy as of May 5, 2015. This is the final and approved form of the dissertation. Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations Jelena Perovanovic Dissertation Research Committee: Eric P. Hoffman, Professor of Integrative Systems Biology, Dissertation Director Anamaris Colberg-Poley, Professor of Integrative Systems Biology, Committee Member Robert J. Freishtat, Associate Professor of Pediatrics, Committee Member Vittorio Sartorelli, Senior Investigator, National Institutes of Health, Committee Member ii © Copyright 2015 by Jelena Perovanovic All rights reserved iii Acknowledgments I am deeply indebted to countless individuals for their support and encouragement during the past five years of graduate studies. First and foremost, I would like to express my gratitude to my mentor, Dr. Eric P. Hoffman, for his unwavering support and guidance, and keen attention to my professional development. This Dissertation would not have been possible without the critical input he provided and the engaging environment he created. -

Exploring Profilin
RESEARCH HIGHLIGHTS NEUROTRANSMISSION Exploring profilin DOI: 10.1038/nrn2182 Surprisingly, they found that Pfn2–/– mice, pointing towards a Pfn2–/– mice displayed normal higher vesicle release probability. development and brain anatomy. Immunoprecipitation and EM studies Learning and memory, long-term also indicated that the number of potentiation (LTP) and long-term docked vesicles is approximately 25% depression (LTD) were not impaired higher in Pfn2–/– mice. in Pfn2–/– mice when compared with How does profilin 2 alter vesicle control mice. These results suggest release? When neurotransmitter that profilin 2 is not required for secretion was stimulated in cortical development, synaptic plasticity or synaptosomes from control mice, learning and memory. F-actin levels were significantly The authors noticed, however, increased. This did not occur in that the Pfn2–/– mice behaved synaptosome preparations from differently to control mice. They Pfn2–/– mice. In addition, profilin 2, were hyperactive, and they showed but not profilin 1, was shown to be elevated exploratory and novelty- tightly associated with the WAVE- Profilins are regulators of actin seeking behaviour when placed into complex, a protein complex that is polymerization. Profilin 2 is a new environment. This included implicated in actin dynamics and expressed mainly in neurons, but increased locomotor activity, wall is abundant in synaptosomal prepa- its specific function is unknown. A rearings and rearings in the centre of rations. The authors therefore study by Boyl et al. provides evidence the experimental area. Conversely, suggest that WAVE and profilin 2 that profilin 2 is required for activ- in the familiar environment of their might cooperate to restrict synaptic ity-stimulated actin polymerization home cage, they showed reduced vesicle release. -

Profilin and Formin Constitute a Pacemaker System for Robust Actin
RESEARCH ARTICLE Profilin and formin constitute a pacemaker system for robust actin filament growth Johanna Funk1, Felipe Merino2, Larisa Venkova3, Lina Heydenreich4, Jan Kierfeld4, Pablo Vargas3, Stefan Raunser2, Matthieu Piel3, Peter Bieling1* 1Department of Systemic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 2Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 3Institut Curie UMR144 CNRS, Paris, France; 4Physics Department, TU Dortmund University, Dortmund, Germany Abstract The actin cytoskeleton drives many essential biological processes, from cell morphogenesis to motility. Assembly of functional actin networks requires control over the speed at which actin filaments grow. How this can be achieved at the high and variable levels of soluble actin subunits found in cells is unclear. Here we reconstitute assembly of mammalian, non-muscle actin filaments from physiological concentrations of profilin-actin. We discover that under these conditions, filament growth is limited by profilin dissociating from the filament end and the speed of elongation becomes insensitive to the concentration of soluble subunits. Profilin release can be directly promoted by formin actin polymerases even at saturating profilin-actin concentrations. We demonstrate that mammalian cells indeed operate at the limit to actin filament growth imposed by profilin and formins. Our results reveal how synergy between profilin and formins generates robust filament growth rates that are resilient to changes in the soluble subunit concentration. DOI: https://doi.org/10.7554/eLife.50963.001 *For correspondence: peter.bieling@mpi-dortmund. mpg.de Introduction Competing interests: The Eukaryotic cells move, change their shape and organize their interior through dynamic actin net- authors declare that no works. -

Epigenetic Regulation of Smad2 and Smad3 by Profilin-2 Promotes Lung Cancer Growth and Metastasis
ARTICLE Received 26 May 2014 | Accepted 29 Jul 2015 | Published 10 Sep 2015 DOI: 10.1038/ncomms9230 Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis Yun-Neng Tang1, Wei-Qiao Ding1, Xiao-Jie Guo1, Xin-Wang Yuan1, Dong-Mei Wang1 & Jian-Guo Song1 Altered transforming growth factor-b (TGF-b) signalling has been implicated in tumour development and progression. However, the molecular mechanism behind this alteration is poorly understood. Here we show that profilin-2 (Pfn2) increases Smad2 and Smad3 expression via an epigenetic mechanism, and that profilin-2 and Smad expression correlate with an unfavourable prognosis of lung cancer patients. Profilin-2 overexpression promotes, whereas profilin-2 knockdown drastically reduces, lung cancer growth and metastasis. We show that profilin-2 suppresses the recruitment of HDAC1 to Smad2 and Smad3 promoters by preventing nuclear translocation of HDAC1 through protein–protein interaction at the C terminus of both proteins, leading to the transcriptional activation of Smad2 and Smad3. Increased Smad2 and Smad3 expression enhances TGF-b1-induced EMT and production of the angiogenic factors VEGF and CTGF. These findings reveal a new regulatory mechanism of TGF-b1/Smad signalling, and suggest a potential molecular target for the development of anticancer drugs. 1 State Key Laboratory of Cell Biology, Innovation Center for Cell Signalling Network, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China. Correspondence and requests for materials should be addressed to J.-G.S. (email: [email protected]). NATURE COMMUNICATIONS | 6:8230 | DOI: 10.1038/ncomms9230 | www.nature.com/naturecommunications 1 & 2015 Macmillan Publishers Limited. -

Evaluation of Profilin 1 As a Biomarker in Myocardial Infarction
European Review for Medical and Pharmacological Sciences 2020; 24: 8112-8116 Evaluation of profilin 1 as a biomarker in myocardial infarction 1 2 3 2 1 1 E. PASZEK , K ŻMUDKA , K. PLENS , J. LEGUTKO , T. RAJS , W. ZAJDEL 1Department of Interventional Cardiology, Institute of Cardiology, John Paul II Hospital, Kraków, Poland 2Jagiellonian University Medical College, Institute of Cardiology, Department of Interventional Cardiology, John Paul II Hospital, Kraków, Poland 3KCRI, Kraków, Poland Abstract. – OBJECTIVE: Profilin 1 Pfn1)( is Introduction likely to be involved in atherogenesis and myo- cardial infarction (MI). Clinical data on this sub- Profilin 1 (Pfn1) is a small, ubiquitous protein ject are very limited. The aim of this study was involved in actin cytoskeleton regulation. Recent to search for associations between serum Pfn1 studies1,2 show a probable link between Pfn1 and and a number of parameters in MI patients: symp- tom onset to PCI time (OPT), myocardial necrosis the pathogenesis of atherosclerosis and myocar- markers, thrombolysis in myocardial infarction dial infarction. So far, data on this subject are (TIMI) flow, antiplatelet drugs, heparin adminis- very limited and mostly come from in vitro and tration and typical atherosclerosis risk factors. animal models. The time of ischemia duration in PATIENTS AND METHODS: We included pa- myocardial infarction (MI) is a crucial parameter tients with type 1 MI (according to the Third Uni- for clinical decision-making and patient progno- versal Definition of Myocardial Infarction) who sis3. In a clinical setting a parameter derived from were able to precisely determine the time of symp- ischemia time – the time from symptom onset to tom onset.