Molecular Phylogenetic Relationship Between and Within the Fruit
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape As A
A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape as a Chiropteran Diversity Hotspot in Sumatra Author(s): Joe Chun-Chia Huang, Elly Lestari Jazdzyk, Meyner Nusalawo, Ibnu Maryanto, Maharadatunkamsi, Sigit Wiantoro, and Tigga Kingston Source: Acta Chiropterologica, 16(2):413-449. Published By: Museum and Institute of Zoology, Polish Academy of Sciences DOI: http://dx.doi.org/10.3161/150811014X687369 URL: http://www.bioone.org/doi/full/10.3161/150811014X687369 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Acta Chiropterologica, 16(2): 413–449, 2014 PL ISSN 1508-1109 © Museum and Institute of Zoology PAS doi: 10.3161/150811014X687369 A recent -
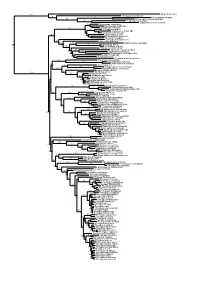
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Index of Handbook of the Mammals of the World. Vol. 9. Bats
Index of Handbook of the Mammals of the World. Vol. 9. Bats A agnella, Kerivoula 901 Anchieta’s Bat 814 aquilus, Glischropus 763 Aba Leaf-nosed Bat 247 aladdin, Pipistrellus pipistrellus 771 Anchieta’s Broad-faced Fruit Bat 94 aquilus, Platyrrhinus 567 Aba Roundleaf Bat 247 alascensis, Myotis lucifugus 927 Anchieta’s Pipistrelle 814 Arabian Barbastelle 861 abae, Hipposideros 247 alaschanicus, Hypsugo 810 anchietae, Plerotes 94 Arabian Horseshoe Bat 296 abae, Rhinolophus fumigatus 290 Alashanian Pipistrelle 810 ancricola, Myotis 957 Arabian Mouse-tailed Bat 164, 170, 176 abbotti, Myotis hasseltii 970 alba, Ectophylla 466, 480, 569 Andaman Horseshoe Bat 314 Arabian Pipistrelle 810 abditum, Megaderma spasma 191 albatus, Myopterus daubentonii 663 Andaman Intermediate Horseshoe Arabian Trident Bat 229 Abo Bat 725, 832 Alberico’s Broad-nosed Bat 565 Bat 321 Arabian Trident Leaf-nosed Bat 229 Abo Butterfly Bat 725, 832 albericoi, Platyrrhinus 565 andamanensis, Rhinolophus 321 arabica, Asellia 229 abramus, Pipistrellus 777 albescens, Myotis 940 Andean Fruit Bat 547 arabicus, Hypsugo 810 abrasus, Cynomops 604, 640 albicollis, Megaerops 64 Andersen’s Bare-backed Fruit Bat 109 arabicus, Rousettus aegyptiacus 87 Abruzzi’s Wrinkle-lipped Bat 645 albipinnis, Taphozous longimanus 353 Andersen’s Flying Fox 158 arabium, Rhinopoma cystops 176 Abyssinian Horseshoe Bat 290 albiventer, Nyctimene 36, 118 Andersen’s Fruit-eating Bat 578 Arafura Large-footed Bat 969 Acerodon albiventris, Noctilio 405, 411 Andersen’s Leaf-nosed Bat 254 Arata Yellow-shouldered Bat 543 Sulawesi 134 albofuscus, Scotoecus 762 Andersen’s Little Fruit-eating Bat 578 Arata-Thomas Yellow-shouldered Talaud 134 alboguttata, Glauconycteris 833 Andersen’s Naked-backed Fruit Bat 109 Bat 543 Acerodon 134 albus, Diclidurus 339, 367 Andersen’s Roundleaf Bat 254 aratathomasi, Sturnira 543 Acerodon mackloti (see A. -

Taxonomy and Natural History of the Southeast Asian Fruit-Bat Genus Dyacopterus
Journal of Mammalogy, 88(2):302–318, 2007 TAXONOMY AND NATURAL HISTORY OF THE SOUTHEAST ASIAN FRUIT-BAT GENUS DYACOPTERUS KRISTOFER M. HELGEN,* DIETER KOCK,RAI KRISTIE SALVE C. GOMEZ,NINA R. INGLE, AND MARTUA H. SINAGA Division of Mammals, National Museum of Natural History (NHB 390, MRC 108), Smithsonian Institution, P.O. Box 37012, Washington, D.C. 20013-7012, USA (KMH) Forschungsinstitut Senckenberg, Senckenberganlage 25, Frankfurt, D-60325, Germany (DK) Philippine Eagle Foundation, VAL Learning Village, Ruby Street, Marfori Heights, Davao City, 8000, Philippines (RKSCG) Department of Natural Resources, Cornell University, Ithaca, NY 14853, USA (NRI) Division of Mammals, Field Museum of Natural History, Chicago, IL 60605, USA (NRI) Museum Zoologicum Bogoriense, Jl. Raya Cibinong Km 46, Cibinong 16911, Indonesia (MHS) The pteropodid genus Dyacopterus Andersen, 1912, comprises several medium-sized fruit-bat species endemic to forested areas of Sundaland and the Philippines. Specimens of Dyacopterus are sparsely represented in collections of world museums, which has hindered resolution of species limits within the genus. Based on our studies of most available museum material, we review the infrageneric taxonomy of Dyacopterus using craniometric and other comparisons. In the past, 2 species have been described—D. spadiceus (Thomas, 1890), described from Borneo and later recorded from the Malay Peninsula, and D. brooksi Thomas, 1920, described from Sumatra. These 2 nominal taxa are often recognized as species or conspecific subspecies representing these respective populations. Our examinations instead suggest that both previously described species of Dyacopterus co-occur on the Sunda Shelf—the smaller-skulled D. spadiceus in peninsular Malaysia, Sumatra, and Borneo, and the larger-skulled D. -

The Evolution of Echolocation in Bats: a Comparative Approach
The evolution of echolocation in bats: a comparative approach Alanna Collen A thesis submitted for the degree of Doctor of Philosophy from the Department of Genetics, Evolution and Environment, University College London. November 2012 Declaration Declaration I, Alanna Collen (née Maltby), confirm that the work presented in this thesis is my own. Where information has been derived from other sources, this is indicated in the thesis, and below: Chapter 1 This chapter is published in the Handbook of Mammalian Vocalisations (Maltby, Jones, & Jones) as a first authored book chapter with Gareth Jones and Kate Jones. Gareth Jones provided the research for the genetics section, and both Kate Jones and Gareth Jones providing comments and edits. Chapter 2 The raw echolocation call recordings in EchoBank were largely made and contributed by members of the ‘Echolocation Call Consortium’ (see full list in Chapter 2). The R code for the diversity maps was provided by Kamran Safi. Custom adjustments were made to the computer program SonoBat by developer Joe Szewczak, Humboldt State University, in order to select echolocation calls for measurement. Chapter 3 The supertree construction process was carried out using Perl scripts developed and provided by Olaf Bininda-Emonds, University of Oldenburg, and the supertree was run and dated by Olaf Bininda-Emonds. The source trees for the Pteropodidae were collected by Imperial College London MSc student Christina Ravinet. Chapter 4 Rob Freckleton, University of Sheffield, and Luke Harmon, University of Idaho, helped with R code implementation. 2 Declaration Chapter 5 Luke Harmon, University of Idaho, helped with R code implementation. Chapter 6 Joseph W. -

Base-Compositional Biases and the Bat Problem. III. the Question of Microchiropteran Monophyly
Base-compositional biases and the bat problem. III. The question of microchiropteran monophyly James M. Hutcheon1,JohnA.W.Kirsch1 and John D. Pettigrew2 1The University of Wisconsin Zoological Museum, 250 North Mills Street, Madison,WI 53706, USA 2Vision,Touch, and Hearing Research Centre, University of Queensland, St Lucia, Queensland 4072, Australia Using single-copy DNA hybridization, we carried out a whole genome study of 16 bats (from ten families) and ¢ve outgroups (two primates and one each dermopteran, scandentian, and marsupial). Three of the bat species represented as many families of Rhinolophoidea, and these always associated with the two representatives of Pteropodidae. All other microchiropterans, however, formed a monophyletic unit displaying interrelationships largely in accord with current opinion. Thus noctilionoids comprised one clade, while vespertilionids, emballonurids, and molossids comprised three others, successively more closely related in that sequence. The unexpected position of rhinolophoids may be due either to the high AT bias they share with pteropodids, or it may be phylogenetically authentic. Reanalysis of the data with varying combinations of the ¢ve outgroups does not indicate a rooting problem, and the inclusion of many bat lineages divided at varying levels similarly discounts long branch attraction as an explanation for the pteropodid^rhinolophoid association. If rhinolophoids are indeed specially related to pteropodids, many synapomorphies of Microchiroptera are called into question, not least the -

Os Nomes Galegos Dos Morcegos 2018 2ª Ed
Os nomes galegos dos morcegos 2018 2ª ed. Citación recomendada / Recommended citation: A Chave (20182): Os nomes galegos dos morcegos. Xinzo de Limia (Ourense): A Chave. http://www.achave.ga"/wp#content/up"oads/achave_osnomes!a"egosdos$morcegos$2018.pd% Para a elaboración deste recurso léxico contouse coa colaboración de Lois de la Calle Carballal. Fotografía: orelludo cincento (Plecotus austriacus ). Autor: Jordi as. &sta o'ra est( su)eita a unha licenza Creative Commons de uso a'erto* con reco+ecemento da autor,a e sen o'ra derivada nin usos comerciais. -esumo da licenza: https://creativecommons.or!/"icences/'.#n #nd//.0/deed.!". Licenza comp"eta: https://creativecommons.or!/"icences/'.#n #nd//.0/"e!a"code0"an!ua!es. 1 !otas introdutorias " que contén este documento Na primeira edición deste documento (2016) fornecéronse denominacións galegas para as especies de morcegos : todas as que están presentes na Galiza (cando menos 2!) e máis de 500 especies do resto do mundo# Nesta segunda edición (2018) incorpórase o logo da %&a'e ao deseño do documento engádese algún nome galego máis e reescr*+ense as notas introdutorias# ,áis completa que a anterior nesta no'a edición ac&éganse nomes galegos para un total de 552 especies# A estrutura En primeiro lugar preséntase unha clasificación taxonómica que considera as superfamilias e familias de morcegos onde se apunta de maneira xeral os nomes dos morcegos que &ai en cada familia# / seguir 'én o corpo do documento unha listaxe onde se indica especie por especie, alén do nome cient*fico os nomes -

Diversity and Diversification Across the Global Radiation of Extant Bats
Diversity and Diversification Across the Global Radiation of Extant Bats by Jeff J. Shi A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Ecology and Evolutionary Biology) in the University of Michigan 2018 Doctoral Committee: Professor Catherine Badgley, co-chair Assistant Professor and Assistant Curator Daniel Rabosky, co-chair Associate Professor Geoffrey Gerstner Associate Research Scientist Miriam Zelditch Kalong (Malay, traditional) Pteropus vampyrus (Linnaeus, 1758) Illustration by Gustav Mützel (Brehms Tierleben), 19271 1 Reproduced as a work in the public domain of the United States of America; accessible via the Wikimedia Commons repository. EPIGRAPHS “...one had to know the initial and final states to meet that goal; one needed knowledge of the effects before the causes could be initiated.” Ted Chiang; Story of Your Life (1998) “Dr. Eleven: What was it like for you, at the end? Captain Lonagan: It was exactly like waking up from a dream.” Emily St. John Mandel; Station Eleven (2014) Bill Watterson; Calvin & Hobbes (October 27, 1989)2 2 Reproduced according to the educational usage policies of, and direct correspondence with Andrews McMeel Syndication. © Jeff J. Shi 2018 [email protected] ORCID: 0000-0002-8529-7100 DEDICATION To the memory and life of Samantha Jade Wang. ii ACKNOWLEDGMENTS All of the research presented here was supported by a National Science Foundation (NSF) Graduate Research Fellowship, an Edwin H. Edwards Scholarship in Biology, and awards from the University of Michigan’s Rackham Graduate School and the Department of Ecology & Evolutionary Biology (EEB). A significant amount of computational work was funded by a Michigan Institute for Computational Discovery and Engineering fellowship; specimen scanning, loans, and research assistants were funded by the Museum of Zoology’s Hinsdale & Walker fund and an NSF Doctoral Dissertation Improvement Grant. -

New Species Records from Buton Island, South East Sulawesi, Including Regional Range Extensions
Melissa Donnelly, Thomas E. Martin, Olivia Cropper, Ellena Yusti, Arthur Arfian, Rachael Smethurst, Catherine Fox, Moira Pryde, Hafirun, Josh Phangurha, Rianne N. van der Aar, Amy Hutchison, Ady Karya, Kangkuso Analuddin, Samsudin, Stephanie K. Courtney Jones Journal of www.secemu.org Bat Research & Conservation DOI: 10.14709/BarbJ.14.1.2021.03 ORIGINAL ARTICLE New species records from Buton Island, South East Sulawesi, including regional range extensions Melissa Donnelly1,2,*, Thomas E. Martin1, Olivia Cropper1,3, Ellena Yusti1, Arthur Arfian1, Rachael Smethurst1, Catherine Fox1,4, Moira Pryde1,5, Hafirun6, Josh Phangurha1, Rianne N. van der Aar1, Amy Hutchison1,7, Ady Karya8, Kangkuso Analuddin8, Samsudin1, Stephanie K. Courtney Jones9,10 1 Operation Wallacea Ltd, Wallace House, Old Bolingbroke, Spilsby, Lincolnshire, ABSTRACT PE23 4EX, UK Peninsular Malaysia is currently thought to host the highest biodiversity of Old World 2 Proyecto CUBABAT, Matanzas, 40100, bats of any region, with 110 species recorded. However, the availability of literature Cuba to facilitate a similarly thorough species ‘checklist’ is not as readily available for 3 Ove Arup & Partners, Leeds, West other parts of Southeast Asia, including Sulawesi, Indonesia. Here we highlight 13 Yorkshire, LS9 8EE, UK new species records from the long-term bat monitoring programme on Buton Island, 4 Long Beach City College, Long Beach, South East Sulawesi, expanding on Patterson et al.’s (2017) previous inventory for this CA 90808, USA study area. One species (Hipposideros galeritus) is a new record for Sulawesi, and seven species (Cynopterus c.f. minutus, Rousettus celebensis, Megaderma spasma, 5 Department of Conservation, Nelson, 7010, New Zealand Hipposideros c.f. -

Teeling2009chap78.Pdf
Bats (Chiroptera) Emma C. Teeling oldest bat fossils (~55 Ma) and is considered a microbat; UCD School of Biology and Environmental Science, Science Center however, the majority of the bat fossil record is fragmen- West, University College Dublin, Belfi eld, Dublin 4, Ireland (emma. tary and missing key species (6, 7). Here I review the rela- [email protected]) tionships and divergence times of the extant families of bats. Abstract Traditionally bats have been divided into two super- ordinal groups: Megachiroptera and Microchiroptera Bats are grouped into 17–18 families (>1000 species) within (see 8, 9 for reviews). Megachiroptera was consid- the mammalian Order Chiroptera. Recent phylogenetic ered basal and contained the Old World megabat fam- analyses of molecular data have reclassifi ed Chiroptera at ily Pteropodidae, whereas Microchiroptera contained the interfamilial level. Traditionally, the non-echolocating the 17 microbat families (8, 9). Although this division megabats (Pteropodidae) have been considered to be the was based mainly on morphological and paleonto- earliest diverging lineage of living bats; however, they are logical data, it highlighted the diB erence in mode of now found to be the closest relatives of the echolocating sensory perception between megabats and microbats. rhinolophoid microbats. Four major groups of echolocating Because all microbats are capable of sophisticated laryn- microbats are supported: rhinolophoids, emballonuroids, geal echolocation whereas megabats are not (5), it was vespertilionoids, and noctilionoids. The timetree suggests believed that laryngeal echolocation had a single origin that the earliest divergences among bats occurred ~64 in the lineage leading to microbats (10). 7 e 17 families million years ago (Ma) and that the four major microbat of microbats have been subsequently divided into two lineages were established by 50 Ma. -

Predicting Wildlife Hosts of Betacoronaviruses for SARS-Cov-2 Sampling Prioritization 2 3 Daniel J
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.22.111344; this version posted May 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 Predicting wildlife hosts of betacoronaviruses for SARS-CoV-2 sampling prioritization 2 3 Daniel J. Becker1,♰, Gregory F. Albery2,♰, Anna R. Sjodin3, Timothée Poisot4, Tad A. Dallas5, Evan 4 A. Eskew6,7, Maxwell J. Farrell8, Sarah Guth9, Barbara A. Han10, Nancy B. Simmons11, and Colin J. 5 Carlson12,13,* 6 7 8 9 ♰ These authors share lead author status 10 * Corresponding author: [email protected] 11 12 1. Department of Biology, Indiana University, Bloomington, IN, U.S.A. 13 2. Department of Biology, Georgetown University, Washington, D.C., U.S.A. 14 3. Department of Biological Sciences, University of Idaho, Moscow, ID, U.S.A. 15 4. Université de Montréal, Département de Sciences Biologiques, Montréal, QC, Canada. 16 5. Department of Biological Sciences, Louisiana State University, Baton Rouge, LA, U.S.A. 17 6. Department of Ecology, Evolution, and Natural Resources, Rutgers University, New Brunswick, 18 NJ, U.S.A. 19 7. Department of Biology, Pacific Lutheran University, Tacoma, WA, U.S.A. 20 8. Department of Ecology & Evolutionary Biology, University of Toronto, Toronto, ON, Canada. 21 9. Department of Integrative Biology, University of California Berkeley, Berkeley, CA, U.S.A. 22 10. Cary Institute of Ecosystem Studies, Millbrook, NY, U.S.A. -

Biogeography and Phylogeny of the Eutheria
FAUNA of AUSTRALIA 36. BIOGEOGRAPHY AND PHYLOGENY OF EUTHERIA G.M. MCKAY, J.H. CALABY & L.S. HALL 1 36. BIOGEOGRAPHY AND PHYLOGENY OF EUTHERIA 2 36. BIOGEOGRAPHY AND PHYLOGENY OF EUTHERIA INTRODUCTION In considering the biogeography and phylogeny of the eutherian mammals in Australia, a distinction must be made between those groups which invaded the continent prior to the 18th Century and those that have been deliberately or inadvertently introduced since then. For the latter group, the worldwide distribution and fossil history is essentially irrelevant except that many species have a long association with humans as either domesticated animals or commensals. In this chapter, we will address the more general questions of the origins and affinities of the native Australian eutherians. For details of the biogeography, fossil history and affinities of the introduced mammals, the reader should refer to the family accounts (Chapters 44–46, 54, 55, 58–62). Any examination of the phylogeny of a group must consider two main lines of evidence: the fossil record and the study of living forms. Morphological studies can be made on both living and fossil forms and such studies, particularly of skeletal and dental characters, have traditionally formed the basis for phylogenetic reconstruction in the Mammalia. In recent years there has been an increase in biochemical and molecular studies on living species which, with a very few exceptions, cannot be compared to extinct species. To date, only a relatively few species of living mammals have been studied adequately using molecular techniques, but attempts are being made to reconcile the biochemical data with the traditional morphological studies (Shoshani 1986; McKenna 1987).