Diversity and Diversification Across the Global Radiation of Extant Bats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
First Records of 10 Bat Species in Guyana and Comments on Diversity of Bats in Iwokrama Forest
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by KU ScholarWorks Acta Chiropterologica, l(2): 179-190,1999 PL ISSN 1508-1 109 O Museum and Institute of Zoology PAS First records of 10 bat species in Guyana and comments on diversity of bats in Iwokrama Forest BURTONK. LIM', MARKD. ENGSTROM~,ROBERT M. TIMM~,ROBERT P. ANDERSON~, and L. CYNTHIAWATSON~ 'Centre for Biodiversity and Conservation Biology, Royal Ontario Museum, 100 Queen's Park, Toronto, Ontario M5S 2C6, Canada; E-mail: [email protected] 2Natural History Museum and Department of Ecology & Evolutionary Biology, University of Kansas, Lawrence, Kansas 66045-2454, USA 3Centrefor the Study of Biological Diversity, University of Guyana, Turkeyen Campus, East Coast Demerara, Guyana Ten species of bats (Centronycteris-maximiliani,Diclidurus albus, D. ingens, D. isabellus, Peropteryx leucoptera, Micronycteris brosseti, M. microtis, Tonatia carrikeri, Lasiurus atratus, and Myotis riparius) collected in the Iwokrarna International Rain Forest Programme site represent the first records of these taxa from Guyana. This report brings the known bat fauna of Guyana to 107 species and the fauna of Iwokrama Forest to 74 species. Measurements, reproductive data, and comments on taxonomy and distribution are provided. Key words: Chiroptera, Neotropics, Guyana, Iwokrama Forest, inventory, species diversity on the first of two field trips that constituted the mammal portion of the faunal survey for The mammalian fauna of Guyana is Iwokrama Forest coordinated through The poorly documented in comparison with Academy of Natural Sciences of Philadel- neighbouring countries in northern South phia. Records from previously unreported America. Most of its species and their distri- specimens at the Royal Ontario Museum are butions are inferred (e.g., Eisenberg, 1989) also presented to augment distributional data. -

Bat Conservation 2021
Bat Conservation Global evidence for the effects of interventions 2021 Edition Anna Berthinussen, Olivia C. Richardson & John D. Altringham Conservation Evidence Series Synopses 2 © 2021 William J. Sutherland This document should be cited as: Berthinussen, A., Richardson O.C. and Altringham J.D. (2021) Bat Conservation: Global Evidence for the Effects of Interventions. Conservation Evidence Series Synopses. University of Cambridge, Cambridge, UK. Cover image: Leucistic lesser horseshoe bat Rhinolophus hipposideros hibernating in a former water mill, Wales, UK. Credit: Thomas Kitching Digital material and resources associated with this synopsis are available at https://www.conservationevidence.com/ 3 Contents Advisory Board.................................................................................... 11 About the authors ............................................................................... 12 Acknowledgements ............................................................................. 13 1. About this book ........................................................... 14 1.1 The Conservation Evidence project ................................................................................. 14 1.2 The purpose of Conservation Evidence synopses ............................................................ 14 1.3 Who this synopsis is for ................................................................................................... 15 1.4 Background ..................................................................................................................... -

Living with Wildlife - Flying Foxes Fact Sheet No
LIVING WITH WILDLIFE - FLYING FOXES FACT SHEET NO. 0063 Living with wildlife - Flying Foxes What are flying foxes? flying foxes use to mark their territory and to attract females during the mating season. Flying foxes, also known as fruit bats, are winged mammals belonging to the sub-order group of megabats. Unlike the smaller insectivorous microbats, the Do flying foxes carry diseases? animal navigates using their eye sight and smell, as opposed to echolocation, Like most wildlife and pets, flying foxes may carry diseases that can affect and feed on nectar, pollen and fruit. Flying foxes forage from over 100 humans. Australian Bat Lyssavirus can be transmitted directly from flying species of native plants and may supplement this diet with introduced plants foxes to humans. The risk of contracting Lyssavirus is extremely low, with found in gardens, orchards and urban areas. transmission only possible through direct contact of saliva from an infected Of the four species of flying foxes native to mainland Australia, three reside animal with a skin penetrating bite or scratch. in the Gladstone Region. These species include the grey-headed flying fox Flying foxes are natural hosts of the Hendra virus, however, there is no (Pteropus poliocephalus), black flying fox (Pteropus alecto) and little red evidence that the virus can be transmitted directly to humans. It is believed flying fox (Pteropus scapulatus). that the virus is transmitted from flying foxes to horses through exposure to All of these species are protected under the Nature Conservation Act urine or birthing fluids. Vaccination is the most effective way of reducing the 1992 and the grey-headed flying fox is also listed as ‘vulnerable’ under the risk of the virus infecting horses. -

Redalyc.Nidification of Polybia Rejecta (Hymenoptera: Vespidae)
Biota Neotropica ISSN: 1676-0611 [email protected] Instituto Virtual da Biodiversidade Brasil Magalhães de Souza, Marcos; Porfiro Pires, Epifânio; Prezoto, Fábio Nidification of Polybia rejecta (Hymenoptera: Vespidae) associated to Azteca chartifex (Hymenoptera: Formicidae) in a fragment of Atlantic Forest, in the state of Minas Gerais, southeastern Brazil Biota Neotropica, vol. 13, núm. 3, julio-septiembre, 2013, pp. 390-392 Instituto Virtual da Biodiversidade Campinas, Brasil Available in: http://www.redalyc.org/articulo.oa?id=199128991038 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Biota Neotrop., vol. 13, no. 3 Nidification of Polybia rejecta (Hymenoptera: Vespidae) associated to Azteca chartifex (Hymenoptera: Formicidae) in a fragment of Atlantic Forest, in the state of Minas Gerais, southeastern Brazil Marcos Magalhães de Souza1, Epifânio Porfiro Pires2,4 & Fábio Prezoto3 1Instituto Federal de Educação, Ciência e Tecnologia do Sul de Minas – IFSULDEMINAS, Campus Inconfidentes, CEP 37576-000, Inconfidentes, MG, Brazil 2Departamento de Entomologia, Universidade Federal de Lavras – UFLA, CEP 37200-000, Lavras, MG, Brazil 3Departamento de Zoologia, Universidade Federal de Juiz de Fora – UFJF, CEP 36036-900, Juiz de Fora, MG, Brazil 4Corresponding author: Epifânio Porfiro Pires, e-mail: [email protected] -

Genetic Diversity of the Chaerephon Leucogaster/Pumilus Complex From
Genetic diversity of the Chaerephon leucogaster/pumilus complex from mainland Africa and the western Indian Ocean islands Theshnie Naidoo 202513500 Submitted in fulfillment of the academic Requirements for the degree of Doctor of Philosophy in the School of Life Sciences, Westville Campus, University of KwaZulu – Natal, Durban. NOVEMBER 2013 Supervisory Committee Prof. JM. Lamb Dr. MC. Schoeman Dr. PJ. Taylor Dr. SM. Goodman i ABSTRACT Chaerephon (Dobson, 1874), an Old World genus belonging to the family Molossidae, is part of the suborder Vespertilioniformes. Members of this genus are distributed across mainland Africa (sample sites; Tanzania, Yemen, Kenya, Botswana, South Africa and Swaziland), its offshore islands (Zanzibar, Pemba and Mozambique Island), Madagascar and the surrounding western Indian Ocean islands (Anjouan, Mayotte, Moheli, Grande Comore, Aldabra and La Reunion). A multifaceted approach was used to elucidate the phylogenetic and population genetic relationships at varying levels amongst these different taxa. Working at the subspecific level, I analysed the phylogenetics and phylogeography of Chaerephon leucogaster from Madagascar, based on mitochondrial cytochrome b and control region sequences. Cytochrome b genetic distances among C. leucogaster samples were low (maximum 0.35 %). Genetic distances between C. leucogaster and C. atsinanana ranged from 1.77 % to 2.62 %. Together, phylogenetic and distance analyses supported the classification of C. leucogaster as a separate species. D-loop data for C. leucogaster samples revealed significant but shallow phylogeographic structuring into three latitudinal groups (13º S, 15 - 17º S, 22 - 23º S) showing exclusive haplotypes which correlated with regions of suitable habitat defined by ecological niche modelling. Population genetic analysis of D-loop sequences indicated that populations from Madagascar have been expanding since 5 842 - 11 143 years BP. -

Daytime Behaviour of the Grey-Headed Flying Fox Pteropus Poliocephalus Temminck (Pteropodidae: Megachiroptera) at an Autumn/Winter Roost
DAYTIME BEHAVIOUR OF THE GREY-HEADED FLYING FOX PTEROPUS POLIOCEPHALUS TEMMINCK (PTEROPODIDAE: MEGACHIROPTERA) AT AN AUTUMN/WINTER ROOST K.A. CONNELL, U. MUNRO AND F.R. TORPY Connell KA, Munro U and Torpy FR, 2006. Daytime behaviour of the grey-headed flying fox Pteropus poliocephalus Temminck (Pteropodidae: Megachiroptera) at an autumn/winter roost. Australian Mammalogy 28: 7-14. The grey-headed flying fox (Pteropus poliocephalus Temminck) is a threatened large fruit bat endemic to Australia. It roosts in large colonies in rainforest patches, mangroves, open forest, riparian woodland and, as native habitat is reduced, increasingly in vegetation within urban environments. The general biology, ecology and behaviour of this bat remain largely unknown, which makes it difficult to effectively monitor, protect and manage this species. The current study provides baseline information on the daytime behaviour of P. poliocephalus in an autumn/winter roost in urban Sydney, Australia, between April and August 2003. The most common daytime behaviours expressed by the flying foxes were sleeping (most common), grooming, mating/courtship, and wing spreading (least common). Behaviours differed significantly between times of day and seasons (autumn and winter). Active behaviours (i.e., grooming, mating/courtship, wing spreading) occurred mainly in the morning, while sleeping predominated in the afternoon. Mating/courtship and wing spreading were significantly higher in April (reproductive period) than in winter (non-reproductive period). Grooming was the only behaviour that showed no significant variation between sample periods. These results provide important baseline data for future comparative studies on the behaviours of flying foxes from urban and ‘natural’ camps, and the development of management strategies for this species. -

Karubats Niouz, Nous Traitons D’Un Sujet D’Actualité En Guade- Les Chauves-Souris Loupe : Le Développement De L’Énergie Éolienne
K arubats Niouz La lettre d’information du Groupe Chiroptères de Guadeloupe N°2 - 2015 Eolien et Chiroptères p.2 Edito Dans ce nouveau numéro de Karubats Niouz, nous traitons d’un sujet d’actualité en Guade- Les chauves-souris loupe : le développement de l’énergie éolienne. en entreprise Nous éclairons le lecteur sur l’impact que peut p.12 avoir cette activité sur certaines espèces de chauves-souris quand leur présence n’est pas suFFisamment prise en compte. Suivi d’une colonie La rubrique "des chauves-souris et des De Fers-de-lances communs hommes » met en exergue un bel exemple de p.23 donnant-donnant entre les Fers-de-lances com- muns et une entreprise locale, Phytobôkaz ! Et notre reporter Manzel Ardops de Fond dupré des Grands-Fonds interroge son Fondateur, le Dr Henri Joseph. Sommaire L’espèce poto mitan de notre biodiversité est le Sturnire de Guadeloupe. Vous découvrirez en 2 Eolien et chiroptères quoi elle participe à la luxuriance de la Forêt tro- 11 Des Chauves-Souris et des Hommes picale humide. Mise en ligne des fiches ‘Molosses‘. L’aide précieuse des chauves-souris au sein de l’entreprise Le zoom espèce est consacré à une espèce Phytobocaz rare, le Chiroderme de la Guadeloupe. 14 Les Chauves-Souris Poto Mitan de notre biodiversité Vous trouverez les synthèses de nos principales . Sturnire de Guadeloupe et ailes à mouches études menées en 2014 et 2015, de l’étude 16 Zoom Espèce : Le Chiroderme de Guadeloupe d’envergure réalisée pour l’évaluation des 18 Etudes et suivis chauves-souris dans les bananeraies aux suivis . -

Predictors of Bat Species Richness Within the Islands of the Caribbean Basin
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Mammalogy Papers: University of Nebraska State Museum Museum, University of Nebraska State 10-11-2019 Predictors of Bat Species Richness within the Islands of the Caribbean Basin Justin D. Hoffman McNeese State University, [email protected] Gabrielle Kadlubar McNeese State University, [email protected] Scott C. Pedersen South Dakota State University, [email protected] Roxanne J. Larsen University of Minnesota, [email protected] Peter A. Larsen University of Minnesota, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/museummammalogy Part of the Biodiversity Commons, Other Ecology and Evolutionary Biology Commons, Terrestrial and Aquatic Ecology Commons, and the Zoology Commons Hoffman, Justin D.; Kadlubar, Gabrielle; Pedersen, Scott C.; Larsen, Roxanne J.; Larsen, Peter A.; Phillips, Carleton J.; Kwiecinski, Gary G.; and Genoways, Hugh H., "Predictors of Bat Species Richness within the Islands of the Caribbean Basin" (2019). Mammalogy Papers: University of Nebraska State Museum. 293. https://digitalcommons.unl.edu/museummammalogy/293 This Article is brought to you for free and open access by the Museum, University of Nebraska State at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Mammalogy Papers: University of Nebraska State Museum by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Justin D. Hoffman, Gabrielle Kadlubar, Scott C. Pedersen, Roxanne J. Larsen, Peter A. Larsen, Carleton J. Phillips, Gary G. Kwiecinski, and Hugh H. Genoways This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ museummammalogy/293 Hoffman, Kadlubar, Pedersen, Larsen, Larsen, Phillips, Kwiecinski, and Genoways in Special Publications / Museum of Texas Tech University, no. -

Bats of Nepal a Field Guide/ /Bats of Nepal a Field Guide
Bats of Nepal A field guide/ /Bats of Nepal A field guide This Publication is supported by: Critical Ecosystem Partnership Fund (CEPF) - World Wildlife Fund WWF Nepal Designed and published by: Small Mammals Conservation and Research Foundation (SMCRF) Compiled and edited by: Pushpa Raj Acharya, Hari Adhikari, Sagar Dahal, Arjun Thapa and Sanjan Thapa Cover photographs: Front cover: Myotis sicarius Mandelli's Mouse-eared Myotis by Sanjan Thapa Back cover: Myotis csorbai Csorba's Mouse-eared Myotis by Sanjan Thapa Cover design: Rajesh Goit First edition 2010 500 copies ISBN 978-9937-2-2951-7 Copyright © 2010 all rights reserved at authors and SMCRF No part of this publication may be reproduced, stored or copied in any form-printed, electronic and photocopied without the written Bats of Nepal permission from the publisher. A field guide Bats of Nepal A field guide/ /Bats of Nepal A field guide forts which strategically put their attention to bat research though we were less experienced and trained. Meanwhile, Bat researches were simultaneously PREFACE supported by international agencies: Bat Conservation International, Lubee Bat Conservancy, Rufford small grants and Chester Zoo. Inconsistent database advocates around 60 species of bat hosted to Nepalese land- scape. Our knowledge on bat fauna is merely based on opportunistic and rare A picture can speak thousand words, we have tried to include maximum pho- effort carried out by foreign scholars bounded with countries biological policy. tographs of the species (about 40 photographs); Most of the bat pictures used in Almost 40 years of biodiversity effort of Nepal, Small mammals has got no re- this book were clicked during different field studies in Nepal. -
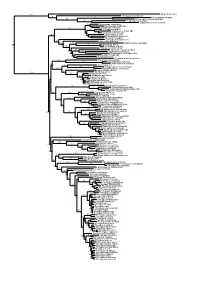
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Chiroptera: Vespertilionidae) from Taiwan and Adjacent China
Zootaxa 3920 (1): 301–342 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2015 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3920.2.6 http://zoobank.org/urn:lsid:zoobank.org:pub:8B991675-0C48-40D4-87D2-DACA524D17C2 Molecular phylogeny and morphological revision of Myotis bats (Chiroptera: Vespertilionidae) from Taiwan and adjacent China MANUEL RUEDI1,5, GÁBOR CSORBA2, LIANG- KONG LIN3 & CHENG-HAN CHOU3,4 1Department of Mammalogy and Ornithology, Natural History Museum of Geneva, Route de Malagnou 1, BP 6434, 1211 Geneva (6), Switzerland. E-mail: [email protected] 2Department of Zoology, Hungarian Natural History Museum, Budapest, Baross u. 13., H-1088. E-mail: [email protected] 3Laboratory of Wildlife Ecology, Department of Biology, Tunghai University, Taichung, Taiwan 407, R.O.C. E-mail: [email protected] 4Division of Zoology, Endemic Species Research Institute, Nantou, Taiwan 552, R.O.C. E-mail: [email protected] 5Corresponding author Table of contents Abstract . 301 Introduction . 302 Material and methods . 310 Results . 314 Discussion . 319 Systematic account . 319 Submyotodon latirostris (Kishida, 1932) . 319 Myotis fimbriatus (Peters, 1870) . 321 Myotis laniger (Peters, 1870) . 322 Myotis secundus sp. n. 324 Myotis soror sp. n. 327 Myotis frater Allen, 1923 . 331 Myotis formosus (Hodgson, 1835) . 334 Myotis rufoniger (Tomes, 1858) . 335 Biogeography and conclusions . 336 Key to the Myotinae from Taiwan and adjacent mainland China . 337 Acknowledgments . 337 References . 338 Abstract In taxonomic accounts, three species of Myotis have been traditionally reported to occur on the island of Taiwan: Watase’s bat (M. -
Siphonaptera)
A peer-reviewed open-access journal ZooKeys 572:Description 7–21 (2016) of two new species of bat fleas of the genusAraeopsylla (Siphonaptera)... 7 doi: 10.3897/zookeys.572.7823 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Description of two new species of bat fleas of the genus Araeopsylla (Siphonaptera) from Kenya and Madagascar with notes on miscellaneous bat fleas Michael W. Hastriter1 1 Monte L. Bean Life Science Museum, Brigham Young University, 290 MLBM, P.O. Box 20200, Provo, Utah 84602-0200, U.S.A. Corresponding author: Michael W. Hastriter ([email protected]) Academic editor: T. Galloway | Received 19 January 2016 | Accepted 25 February 2016 | Published 15 March 2016 http://zoobank.org/2322CF20-1A94-4A0D-BE3F-F8212E737812 Citation: Hastriter MW (2016) Description of two new species of bat fleas of the genus Araeopsylla (Siphonaptera) from Kenya and Madagascar with notes on miscellaneous bat fleas. ZooKeys 572: 7–21.doi: 10.3897/zookeys.572.7823 Abstract The flea genus Araeopsylla Jordan and Rothschild, 1921 contains nine species distributed throughout the Palaearctic, Ethiopian and Oriental Regions primarily on mollosid bats. A new species of bat flea, Araeopsylla goodmani, is described. This new species is represented by three females collected from one male specimen of the mollosid bat Chaerephon jobimena Goodman & Cardiff, 2004 from Fianarantsoa Province, Madagascar. A second new species, Araeopsylla smiti, is described from one male from the Rift Valley, Kenya. It was collected from the molossid bat Chaerephon bivittatus (Heuglin, 1861). This rep- resents the first record of Araeopsylla in Kenya. Previous records of Araeopsylla in the Malagasy region included Araeopsylla martialis (Rothschild, 1903) from Reunion Island and Madagascar.