The Evolution of Echolocation in Bats: a Comparative Approach
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bat Calls of New South Wales
Bat calls of New South Wales Region based guide to the echolocation calls of microchiropteran bats Michael Pennay1 , Brad Law2 & Linda Reinhold3 1 New South Wales Department of Environment and Conservation 2 State Forests of New South Wales 3 Queensland Department of Natural Resources and Mines Bat calls of New South Wales Bat calls of New South Wales Published by the NSW Department of Environment and Conservation May 2004 Copyright © NSW Department of Environment and Conservation ISBN 0 7313 6786 3 This guide is the result of a co-operative project between NSW National Parks and Wildlife Service, now the NSW Department of Environment and Conservation (DEC) and State Forests of NSW (SFNSW). DEC provided project funding, management, staff, reference calls, preparation and printing. SFNSW provided part funding and granted support of staff time and expertise, reference calls and editing. Research was conducted under NPWS scientific licence number A2753 and SFNSW special purpose permit for research number 05466. Material presented in this publication may be copied for personal use or republished for non-commercial purposes provided that NSW Department of Environment and Conservation is fully acknowledged as the copyright owner. Apart from these purposes or for private study, research, criticism or review, as permitted under the Australian Copyright Act, no part of this publication may be reproduced by any process without written permission from NSW Department of Environment and Conservation. Inquiries should be addressed to the NSW Department of Environment and Conservation. This publication should be cited as follows: Pennay, M., Law, B., Reinhold, L. (2004). Bat calls of New South Wales: Region based guide to the echolocation calls of Microchiropteran bats. -

Bat Conservation 2021
Bat Conservation Global evidence for the effects of interventions 2021 Edition Anna Berthinussen, Olivia C. Richardson & John D. Altringham Conservation Evidence Series Synopses 2 © 2021 William J. Sutherland This document should be cited as: Berthinussen, A., Richardson O.C. and Altringham J.D. (2021) Bat Conservation: Global Evidence for the Effects of Interventions. Conservation Evidence Series Synopses. University of Cambridge, Cambridge, UK. Cover image: Leucistic lesser horseshoe bat Rhinolophus hipposideros hibernating in a former water mill, Wales, UK. Credit: Thomas Kitching Digital material and resources associated with this synopsis are available at https://www.conservationevidence.com/ 3 Contents Advisory Board.................................................................................... 11 About the authors ............................................................................... 12 Acknowledgements ............................................................................. 13 1. About this book ........................................................... 14 1.1 The Conservation Evidence project ................................................................................. 14 1.2 The purpose of Conservation Evidence synopses ............................................................ 14 1.3 Who this synopsis is for ................................................................................................... 15 1.4 Background ..................................................................................................................... -

Volume 41, 2000
BAT RESEARCH NEWS Volume 41 : No. 1 Spring 2000 I I BAT RESEARCH NEWS Volume 41: Numbers 1–4 2000 Original Issues Compiled by Dr. G. Roy Horst, Publisher and Managing Editor of Bat Research News, 2000. Copyright 2011 Bat Research News. All rights reserved. This material is protected by copyright and may not be reproduced, transmitted, posted on a Web site or a listserve, or disseminated in any form or by any means without prior written permission from the Publisher, Dr. Margaret A. Griffiths. The material is for individual use only. Bat Research News is ISSN # 0005-6227. BAT RESEARCH NEWS Volume41 Spring 2000 Numberl Contents Resolution on Rabies Exposure Merlin Tuttle and Thomas Griffiths o o o o eo o o o • o o o o o o o o o o o o o o o o 0 o o o o o o o o o o o 0 o o o 1 E - Mail Directory - 2000 Compiled by Roy Horst •••• 0 ...................... 0 ••••••••••••••••••••••• 2 ,t:.'. Recent Literature Compiled by :Margaret Griffiths . : ....••... •"r''• ..., .... >.•••••• , ••••• • ••< ...... 19 ,.!,..j,..,' ""o: ,II ,' f 'lf.,·,,- .,'b'l: ,~··.,., lfl!t • 0'( Titles Presented at the 7th Bat Researc:b Confei'ebee~;Moscow :i'\prill4-16~ '1999,., ..,, ~ .• , ' ' • I"',.., .. ' ""!' ,. Compiled by Roy Horst .. : .......... ~ ... ~· ....... : :· ,"'·~ .• ~:• .... ; •. ,·~ •.•, .. , ........ 22 ·.t.'t, J .,•• ~~ Letters to the Editor 26 I ••• 0 ••••• 0 •••••••••••• 0 ••••••• 0. 0. 0 0 ••••••• 0 •• 0. 0 •••••••• 0 ••••••••• 30 News . " Future Meetings, Conferences and Symposium ..................... ~ ..,•'.: .. ,. ·..; .... 31 Front Cover The illustration of Rhinolophus ferrumequinum on the front cover of this issue is by Philippe Penicaud . from his very handsome series of drawings representing the bats of France. -

Diet and the Evolution of Digestion and Renal Function in Phyllostomid Bats
Zoology 104 (2001): 59–73 © by Urban & Fischer Verlag http://www.urbanfischer.de/journals/zoology Diet and the evolution of digestion and renal function in phyllostomid bats Jorge E. Schondube1,*, L. Gerardo Herrera-M.2 and Carlos Martínez del Rio1 1Department of Ecology and Evolutionary Biology, University of Arizona, Tucson 2Instituto de Biología, Departamento de Zoología, Universidad Nacional Autónoma de México, México Received: April 2, 2001 · Accepted: April 12, 2001 Abstract Bat species in the monophyletic family Phyllostomidae feed on blood, insects, small vertebrates, nectar, fruit and complex omnivorous mixtures. We used nitrogen stable isotope ratios to characterize bat diets and adopted a phylogenetically informed approach to investi- gate the physiological changes that accompany evolutionary diet changes in phyllostomids. We found that nitrogen stable isotopes sep- arated plant-eating from animal-eating species. The blood of the latter was enriched in 15N. A recent phylogenetic hypothesis suggests that with the possible exception of carnivory, which may have evolved twice, all diets evolved only once from insectivory. The shift from insectivory to nectarivory and frugivory was accompanied by increased intestinal sucrase and maltase activity, decreased trehalase activity, and reduced relative medullary thickness of kidneys. The shift from insectivory to sanguinivory and carnivory resulted in re- duced trehalase activity. Vampire bats are the only known vertebrates that do not exhibit intestinal maltase activity. We argue that these physiological changes are adaptive responses to evolutionary diet shifts. Key words: Bats, comparative method, diet, digestive and renal function, stable isotopes. Introduction The family Phyllostomidae is a speciose (49 genera and Characterizing animal diets can be difficult. -

A New Species of the Genus Murina (Chiroptera: Vespertilionidae) from the Central Highlands of Vietnam with a Review of the Subfamily Murininae in Vietnam
Acta Chiropterologica, 17(2): 201–232, 2015 PL ISSN 1508-1109 © Museum and Institute of Zoology PAS doi: 10.3161/15081109ACC2015.17.2.001 A new species of the genus Murina (Chiroptera: Vespertilionidae) from the Central Highlands of Vietnam with a review of the subfamily Murininae in Vietnam NGUYEN TRUONG SON1, 8, GABOR CSORBA2, VUONG TAN TU1, VU DINH THONG1, YI WU3, MASASHI HARADA4, TATSUO OSHIDA5, HIDEKI ENDO6, and MASAHARU MOTOKAWA7 1Institute of Ecology and Biological Resources, Vietnam Academy of Sciences and Technology, 18 Hoang Quoc Viet St., Caugiay, Hanoi, Vietnam 2Department of Zoology, Hungarian Natural History Museum, H1088 Budapest, Baross u.13, Hungary 3College of Life Science, Guangzhou University, Guangzhou 510006, China 4Graduate School of Medicine, Osaka City University, Osaka 545-8585, Japan 5Laboratory of Wildlife Ecology, Obihiro University of Agriculture and Veterinary Medicine, Obihiro 080-8555, Japan 6The University Museum, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan 7The Kyoto University Museum, Kyoto University, Sakyo, Kyoto 606-8501, Japan 8Corresponding author: E-mail: [email protected] The subfamily Murininae has high species diversity in Vietnam, but taxonomic studies are limited. In this paper, we describe a new species of the genus Murina based on a specimen collected from Ngoc Linh Nature Reserve, Kon Tum Province in the Central Highlands of Vietnam. It is a medium-sized species with ‘suilla-type’ dentition. A taxonomic review of Murininae from Vietnam was also conducted based on combined morphological, DNA, and karyological characteristics. Molecular phylogenetic analyses based on the mitochondrial cytochrome c oxidase subunit (COI) gene supported the subfamily Murininae, while the genus Murina proved to be paraphyletic in relation to the genera Harpiocephalus and Harpiola. -

Bats of Nepal a Field Guide/ /Bats of Nepal a Field Guide
Bats of Nepal A field guide/ /Bats of Nepal A field guide This Publication is supported by: Critical Ecosystem Partnership Fund (CEPF) - World Wildlife Fund WWF Nepal Designed and published by: Small Mammals Conservation and Research Foundation (SMCRF) Compiled and edited by: Pushpa Raj Acharya, Hari Adhikari, Sagar Dahal, Arjun Thapa and Sanjan Thapa Cover photographs: Front cover: Myotis sicarius Mandelli's Mouse-eared Myotis by Sanjan Thapa Back cover: Myotis csorbai Csorba's Mouse-eared Myotis by Sanjan Thapa Cover design: Rajesh Goit First edition 2010 500 copies ISBN 978-9937-2-2951-7 Copyright © 2010 all rights reserved at authors and SMCRF No part of this publication may be reproduced, stored or copied in any form-printed, electronic and photocopied without the written Bats of Nepal permission from the publisher. A field guide Bats of Nepal A field guide/ /Bats of Nepal A field guide forts which strategically put their attention to bat research though we were less experienced and trained. Meanwhile, Bat researches were simultaneously PREFACE supported by international agencies: Bat Conservation International, Lubee Bat Conservancy, Rufford small grants and Chester Zoo. Inconsistent database advocates around 60 species of bat hosted to Nepalese land- scape. Our knowledge on bat fauna is merely based on opportunistic and rare A picture can speak thousand words, we have tried to include maximum pho- effort carried out by foreign scholars bounded with countries biological policy. tographs of the species (about 40 photographs); Most of the bat pictures used in Almost 40 years of biodiversity effort of Nepal, Small mammals has got no re- this book were clicked during different field studies in Nepal. -
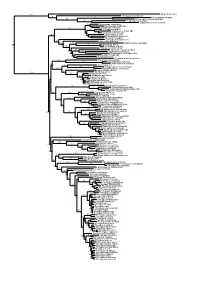
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

The Australasian Bat Society Newsletter
The Australasian Bat Society Newsletter Number 29 November 2007 ABS Website: http://abs.ausbats.org.au ABS Listserver: http://listserv.csu.edu.au/mailman/listinfo/abs ISSN 1448-5877 The Australasian Bat Society Newsletter, Number 29, November 2007 – Instructions for contributors – The Australasian Bat Society Newsletter will accept contributions under one of the following two sections: Research Papers, and all other articles or notes. There are two deadlines each year: 31st March for the April issue, and 31st October for the November issue. The Editor reserves the right to hold over contributions for subsequent issues of the Newsletter, and meeting the deadline is not a guarantee of immediate publication. Opinions expressed in contributions to the Newsletter are the responsibility of the author, and do not necessarily reflect the views of the Australasian Bat Society, its Executive or members. For consistency, the following guidelines should be followed: • Emailed electronic copy of manuscripts or articles, sent as an attachment, is the preferred method of submission. Manuscripts can also be sent on 3½” floppy disk, preferably in IBM format. Please use the Microsoft Word template if you can (available from the editor). Faxed and hard copy manuscripts will be accepted but reluctantly! Please send all submissions to the Newsletter Editor at the email or postal address below. • Electronic copy should be in 11 point Arial font, left and right justified with 16 mm left and right margins. Please use Microsoft Word; any version is acceptable. • Manuscripts should be submitted in clear, concise English and free from typographical and spelling errors. Please leave two spaces after each sentence. -

2020 Special Issue
Journal Home page : www.jeb.co.in « E-mail : [email protected] Original Research Journal of Environmental Biology TM p-ISSN: 0254-8704 e-ISSN: 2394-0379 JEB CODEN: JEBIDP DOI : http://doi.org/10.22438/jeb/4(SI)/MS_1904 Plagiarism Detector Grammarly New records and present status of bat fauna in Mizoram, North-Eastern India C. Vanlalnghaka Department of Zoology, Govt. Serchhip College, Mizoram–796 181, India *Corresponding Author Email : [email protected] Paper received: 08.12.2019 Revised received: 24.06.2020 Accepted: 10.07.2020 Abstract Aim: The present study aimed to investigate the diversity of bat fauna in Mizoram and prepare a checklist for future references. This study also investigated threats and suggested recommendations for implementing conservation measures for bat fauna in Mizoram. Methodology: The present study was carried out in different parts of Mizoram between January 2012 - October 2019. Bats were trapped by using mist nets and hoop nets. Diagnostic morphological characters of bat were used for species identification. Digital camera and video camera were also used for further identification and documentation of bats. Results: During January 2012 – December 2016, eighteen bat species were identified. Recently, from January 2017 - October 2019 insectivorous bat species, Scotomanes ornatus was first time documented in Serchhip District (23.3 ºN 92.83 ºE), Mizoram. In total nineteen bat species were identified in this study, out of which ten species were first time recorded and nine species were rediscovered from the previous documentation. From the previous and present data, total of thirty-six bat Study the diversity of bat fauna and prepared checklist in species were recorded in Mizoram- nine Mizoram. -

Desmodus Rotundus) Blood Feeding
toxins Article Vampire Venom: Vasodilatory Mechanisms of Vampire Bat (Desmodus rotundus) Blood Feeding Rahini Kakumanu 1, Wayne C. Hodgson 1, Ravina Ravi 1, Alejandro Alagon 2, Richard J. Harris 3 , Andreas Brust 4, Paul F. Alewood 4, Barbara K. Kemp-Harper 1,† and Bryan G. Fry 3,*,† 1 Department of Pharmacology, Biomedicine Discovery Institute, Faculty of Medicine, Nursing & Health Sciences, Monash University, Clayton, Victoria 3800, Australia; [email protected] (R.K.); [email protected] (W.C.H.); [email protected] (R.R.); [email protected] (B.K.K.-H.) 2 Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Av. Universidad 2001, Cuernavaca, Morelos 62210, Mexico; [email protected] 3 Venom Evolution Lab, School of Biological Sciences, University of Queensland, St. Lucia, Queensland 4067, Australia; [email protected] 4 Institute for Molecular Biosciences, University of Queensland, St Lucia, QLD 4072, Australia; [email protected] (A.B.); [email protected] (P.F.A.) * Correspondence: [email protected] † Joint senior authors. Received: 20 November 2018; Accepted: 2 January 2019; Published: 8 January 2019 Abstract: Animals that specialise in blood feeding have particular challenges in obtaining their meal, whereby they impair blood hemostasis by promoting anticoagulation and vasodilation in order to facilitate feeding. These convergent selection pressures have been studied in a number of lineages, ranging from fleas to leeches. However, the vampire bat (Desmondus rotundus) is unstudied in regards to potential vasodilatory mechanisms of their feeding secretions (which are a type of venom). This is despite the intense investigations of their anticoagulant properties which have demonstrated that D. -

Occasional Papers Museum of Texas Tech University Number 295 6 July 2010
Occasional Papers Museum of Texas Tech University Number 295 6 July 2010 Karyology of five SpecieS of BatS (veSpertilionidae, HippoSideridae, and nycteridae) from gaBon witH commentS on tHe taxonomy of Glauconycteris Calvin a. Porter, ashley W. Primus, FederiCo G. hoFFmann, and robert J. baker aBStract We karyotyped five species of bats from Gabon. Glauconycteris beatrix and G. poensis both have an all-biarmed 2n = 22 karyotype, consistent with the recognition of Glauconycteris as a genus distinct from Chalinolobus. One specimen of Hipposideros caffer had a 2n = 32 karyotype similar to that published for this species from other areas in Africa. We report a 2n = 52 karyotype for Hipposideros gigas which is identical to that found in H. vittatus. The slit-faced bat Nycteris grandis has a 2n = 42 karyotype similar to that known in other species of Nycteris. Key words: chromosomes, Gabon, Glauconycteris, Hipposideros, karyotypes, Nycteris, Rabi, taxonomy introduction The Republic of Gabon includes extensive tracts documented the presence of 13 chiropteran species in of tropical rain forest and has an economy based the rainforest of the Rabi Oilfield. Primus et al. (2006) largely on oil production. A recent study of biodiversity reported karyotypes of four species of shrews, seven (Alonso et al. 2006; Lee et al. 2006) focused on the species of rodents, and five species of megachiropteran Rabi Oilfield, which is located in the Gamba Complex bats collected at Rabi. However, they did not describe of Protected Areas in the Ogooué-Maritime Province chromosomal data for the microchiropteran specimens of southwestern Gabon. This study included a survey pending confirmation of species identifications. -

Lasiurus Ega (Southern Yellow Bat)
UWI The Online Guide to the Animals of Trinidad and Tobago Ecology Lasiurus ega (Southern Yellow Bat) Family: Vespertilionidae (Vesper or Evening Bats) Order: Chiroptera (Bats) Class: Mammalia (Mammals) Fig. 1. Southern yellow bat, Lasiurus ega. [http://www.collett-trust.org/uploads/Dasypterus%20ega5.jpg, downloaded 16 February 2016] TRAITS. Lasiurus ega is medium sized with relatively long ears and dull yellow/orange fur (Fig. 1). The span of its wing is approximately 35cm. Its mean body length is 11.8cm; tail length, 5.1cm; foot length, 0.90cm and forearm length 4.7cm and its mass ranges from 10-18g; however, the males are smaller in length and size than the females. They have a short body with a lateral projection of the upper lip and short, rounded ears (Fig. 2). The digits shorten from the 3rd to the 5th finger. Each female possesses 4 mammae whereas the males have a distally spiny penis (Kurta and Lehr, 1995). DISTRIBUTION. Found from the southwestern United States to northern Argentina and Uruguay; also can be found in Mexico and Central America (Fig. 3). The southern yellow bat is native to Trinidad. It has a seasonal migratory pattern in which it moves from the north to avoid the harsh/cold conditions (Encyclopedia of Life, 2016). UWI The Online Guide to the Animals of Trinidad and Tobago Ecology HABITAT AND ACTIVITY. The habitat of the southern yellow bat is forest which has a lot of wooded surroundings, foliage as well as palms. The bat is nocturnal in nature. Sometimes it inhabits thatched roofing and corn stalks that are dried, however they avoid entering mountainous areas.