Novel Associations of Nonstructural Loci with Paraoxonase Activity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Silencing of Phosphoinositide-Specific
ANTICANCER RESEARCH 34: 4069-4076 (2014) Silencing of Phosphoinositide-specific Phospholipase C ε Remodulates the Expression of the Phosphoinositide Signal Transduction Pathway in Human Osteosarcoma Cell Lines VINCENZA RITA LO VASCO1, MARTINA LEOPIZZI2, DANIELA STOPPOLONI3 and CARLO DELLA ROCCA2 Departments of 1Sense Organs , 2Medicine and Surgery Sciences and Biotechnologies and 3Biochemistry Sciences “A. Rossi Fanelli”, Sapienza University, Rome, Italy Abstract. Background: Ezrin, a member of the signal transduction pathway (5). The reduction of PIP2 ezrin–radixin–moesin family, is involved in the metastatic induces ezrin dissociation from the plasma membrane (6). spread of osteosarcoma. Ezrin binds phosphatydil inositol-4,5- The levels of PIP2 are regulated by the PI-specific bisphosphate (PIP2), a crucial molecule of the phospholipase C (PI-PLC) family (7), constituting thirteen phosphoinositide signal transduction pathway. PIP2 levels are enzymes divided into six sub-families on the basis of amino regulated by phosphoinositide-specific phospholipase C (PI- acid sequence, domain structure, mechanism of recruitment PLC) enzymes. PI-PLCε isoform, a well-characterized direct and tissue distribution (7-15). PI-PLCε, a direct effector of effector of rat sarcoma (RAS), is at a unique convergence RAS (14-15), might be the point of convergence for the point for the broad range of signaling pathways that promote broad range of signalling pathways that promote the RAS GTPase-mediated signalling. Materials and Methods. By RASGTPase-mediated signalling (16). using molecular biology methods and microscopic analyses, In previous studies, we suggested a relationship between we analyzed the expression of ezrin and PLC genes after PI-PLC expression and ezrin (17-18). -
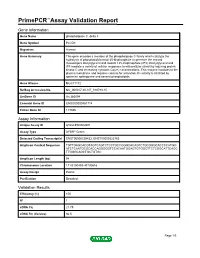
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name phospholipase C, delta 3 Gene Symbol PLCD3 Organism Human Gene Summary This gene encodes a member of the phospholipase C family which catalyze the hydrolysis of phosphatidylinositol 45-bisphosphate to generate the second messengers diacylglycerol and inositol 145-trisphosphate (IP3). Diacylglycerol and IP3 mediate a variety of cellular responses to extracellular stimuli by inducing protein kinase C and increasing cytosolic Ca(2+) concentrations. This enzyme localizes to the plasma membrane and requires calcium for activation. Its activity is inhibited by spermine sphingosine and several phospholipids. Gene Aliases MGC71172 RefSeq Accession No. NC_000017.10, NT_010783.15 UniGene ID Hs.380094 Ensembl Gene ID ENSG00000161714 Entrez Gene ID 113026 Assay Information Unique Assay ID qHsaCED0002601 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000539433, ENST00000322765 Amplicon Context Sequence TGTTGAGCACGTAGTCAGTCTCCTGCCGGGCACAGTCTGCGGGCACCCCATGG ATCTCAATGCGCACCAGGGGGTCCACAATGGAGTGTGGCTTCTCGGCATTCAGC TTGGGCAGCTGCTGTGC Amplicon Length (bp) 94 Chromosome Location 17:43190493-43190616 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 1 cDNA Cq 21.75 cDNA Tm (Celsius) 86.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 23.62 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PLCD3, Human Amplification Plot Amplification of cDNA generated from -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

Transcriptomic Uniqueness and Commonality of the Ion Channels and Transporters in the Four Heart Chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A
www.nature.com/scientificreports OPEN Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A. Iacobas3,4* Myocardium transcriptomes of left and right atria and ventricles from four adult male C57Bl/6j mice were profled with Agilent microarrays to identify the diferences responsible for the distinct functional roles of the four heart chambers. Female mice were not investigated owing to their transcriptome dependence on the estrous cycle phase. Out of the quantifed 16,886 unigenes, 15.76% on the left side and 16.5% on the right side exhibited diferential expression between the atrium and the ventricle, while 5.8% of genes were diferently expressed between the two atria and only 1.2% between the two ventricles. The study revealed also chamber diferences in gene expression control and coordination. We analyzed ion channels and transporters, and genes within the cardiac muscle contraction, oxidative phosphorylation, glycolysis/gluconeogenesis, calcium and adrenergic signaling pathways. Interestingly, while expression of Ank2 oscillates in phase with all 27 quantifed binding partners in the left ventricle, the percentage of in-phase oscillating partners of Ank2 is 15% and 37% in the left and right atria and 74% in the right ventricle. The analysis indicated high interventricular synchrony of the ion channels expressions and the substantially lower synchrony between the two atria and between the atrium and the ventricle from the same side. Starting with crocodilians, the heart pumps the blood through the pulmonary circulation and the systemic cir- culation by the coordinated rhythmic contractions of its upper lef and right atria (LA, RA) and lower lef and right ventricles (LV, RV). -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

PLCD3, a Flotillin2-Interacting Protein, Is Involved in Proliferation, Migration and Invasion of Nasopharyngeal Carcinoma Cells
ONCOLOGY REPORTS 39: 45-52, 2018 PLCD3, a flotillin2-interacting protein, is involved in proliferation, migration and invasion of nasopharyngeal carcinoma cells WeIDoNg LIu1,2*, XuXu LIu1,2*, LeI WANg1,2, BIN ZHu1,2, CHANG ZHANG1,2, WeI JIA1,2, HeCHeNg ZHu3, XINgDoNg LIu3, MeIZuo ZHoNg3, DAN XIE4, YANYu LIu1,2, SHASHA LI1,2, JIA SHI1,2, JIANXINg LIN1,2, XIAOMENG XIA5, XINgJuN JIANg6 and CAIPING REN1,2 1The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health and the Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of education, Xiangya Hospital, Central South university, Changsha, Hunan 410008; 2Cancer Research Institute, Collaborative Innovation Center for Cancer Medicine, School of Basic Medical Science, Central South university, Changsha, Hunan 410078; 3Changsha Kexin Cancer Hospital, Changsha, Hunan 410205; 4State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen university Cancer Center, guangzhou, Hunan 510060; 5Department of Gynecology and Obstetrics, The Second Xiangya Hospital, Central South university, Changsha, Hunan 410011; 6Department of Neurosurgery, Xiangya Hospital, Central South university, Changsha, Hunan 410008, P.R. China Received April 1, 2017; Accepted September 18, 2017 DoI: 10.3892/or.2017.6080 Abstract. Phospholipase C (PLC) is a pivotal enzyme in the tial of 5-8F, a highly metastatic NPC cell line, by restraining phosphoinositide pathway that promotes the second messen- its growth, proliferation, mobility and migration. The present gers, diacylglycerol (DAG) and inositol 1,4,5-trisphosphate study demonstrated that PLCD3 may be an oncogenic protein (IP3), to participate in eukaryotic signal transduction. Several in NPC and that it plays an important role in the progression PLC isozymes are associated with cancer, such as PLC-β1, of NPC partially by interacting with Flot2. -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

UP780, a Chromone-Enriched Aloe Composition, Enhances Adipose Insulin Receptor Signaling and Decreases Liver Lipid Biosynthesis
Open Journal of Genetics, 2013, 3, 9-86 OJGen http://dx.doi.org/10.4236/ojgen.2013.32A2002 Published Online July 2013 (http://www.scirp.org/journal/ojgen/) UP780, a Chromone-Enriched Aloe Composition, Enhances Adipose Insulin Receptor Signaling and Decreases Liver Lipid Biosynthesis Julie Tseng-Crank1*, Seon-Gil Do2, Brandon Corneliusen1, Carmen Hertel1, Jennifer Homan1, Mesfin Yimam1, Jifu Zhao1, Qi Jia1 1Unigen, Seattle, USA 2Unigen Korea, Songjung-Ri, Byeongchen-Myeon, Vheonan-Si, Chungnam, South Korea Email: *[email protected] Received 12 May 2013; revised 17 June 2013; accepted 17 July 2013 Copyright © 2013 Julie Tseng-Crank et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT 1. INTRODUCTION Nutrigenomic studies were conducted to uncover the Diabetes and prediabetes have become epidemic globally. mechanism of action for the hypoglycemic and insulin In recent US surveys, 26.9% of the population aged 65 sensitizing effects of UP780. From high fat diet-induc- years and older had diabetes and 35% of those 20 years ed obesity mouse model for UP780, livers and white or older had prediabetes [1]. WHO of the United Nations adipose tissues (WAT) from groups of lean control, estimated in 2011 that 346 million people worldwide had high fat diet (HFD), and HFD treated with UP780 were diabetes, and that number was likely to double by 2030 collected for microarray study. Microarray genera- [2]. Complications of diabetes include heart disease, ted gene expression changes were applied to Ingenu- stroke, hypertension, blindness, kidney disease, neuro- ity Pathway Analysis for changes in canonical meta- pathy, amputation, and dental disease. -

Differences in Metabolomic and Transcriptomic Profiles Between
Genes Nutr (2013) 8:411–423 DOI 10.1007/s12263-012-0328-0 RESEARCH PAPER Differences in metabolomic and transcriptomic profiles between responders and non-responders to an n-3 polyunsaturated fatty acids (PUFAs) supplementation Iwona Rudkowska • Ann-Marie Paradis • Elisabeth Thifault • Pierre Julien • Olivier Barbier • Patrick Couture • Simone Lemieux • Marie-Claude Vohl Received: 9 July 2012 / Accepted: 27 November 2012 / Published online: 19 December 2012 Ó Springer-Verlag Berlin Heidelberg 2012 Abstract Studies have demonstrated large within-popu- key genes in lipid metabolism: fatty acid desaturase 2, lation heterogeneity in plasma triacylglycerol (TG) phospholipase A2 group IVA, arachidonate 15-lipoxyge- response to n-3 PUFA supplementation. The objective of nase, phosphatidylethanolamine N-methyltransferase, the study was to compare metabolomic and transcriptomic monoglyceride lipase, and glycerol-3-phosphate acyl- profiles of responders and non-responders of an n-3 PUFA transferase, were expressed in opposing direction between supplementation. Thirty subjects completed a 2-week run- subgroups. In sum, results highlight key differences in lipid in period followed by a 6-week supplementation with n-3 metabolism of non-responders compared to responders PUFA (3 g/d). Six subjects did not lower their plasma TG after an n-3 PUFA supplementation, which may explain the (?9 %) levels (non-responders) and were matched to 6 inter-individual variability in plasma TG response. subjects who lowered TG (-41 %) concentrations (responders) after the n-3 PUFA supplementation. Pre-n-3 Keywords Lipidomics Á Metabolic pathways Á PUFA supplementation characteristics did not differ Metabolites Á Microarray Á Nutrigenomics between the non-responders and responders except for plasma glucose concentrations. -

ANGPTL3 Deficiency Alters the Lipid Profile and Metabolism of Cultured Hepatocytes
*REVISED Manuscript (text UNmarked) Click here to view linked References ANGPTL3 deficiency alters the lipid profile and metabolism of cultured hepatocytes and human lipoproteins 1,2,3 1 4 1 Hanna Ruhanen , Nidhina Haridas P.A. , Ilenia Minicocci , Juuso H. Taskinen , Francesco Palmas5, Alessia di Costanzo4, Laura D’Erasmo4, Jari Metso1, Jennimari Partanen1, Jesmond Dalli5,6, You Zhou7, Marcello Arca4, Matti Jauhiainen1, Reijo Käkelä2,3 & Vesa M. Olkkonen1,8,* 1Minerva Foundation Institute for Medical Research, Helsinki, Finland; 2Molecular and Integrative Biosciences, University of Helsinki, Helsinki, Finland; 3Helsinki University Lipidomics Unit (HiLIPID), Helsinki Institute for Life Science (HiLIFE), Helsinki, Finland; 4Department of Translational and Precision Medicine, Sapienza University of Rome, Italy, 5Lipid Mediator Unit, William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom; 6Centre for Inflammation and Therapeutic Innovation, Queen Mary University of London, London, UK; 7Systems Immunity University Research Institute and Division of Infection & Immunity, Cardiff University, Cardiff, United Kingdom, 8Department of Anatomy, University of Helsinki, Finland. *Corresponding author at: Minerva Foundation Institute for Medical Research, Biomedicum 2U, Tukholmankatu 8, FI-00290 Helsinki, Finland; Tel +358-2-94125705, e-mail [email protected] 1 ABSTRACT Loss-of-function (LOF) mutations in ANGPTL3, an inhibitor of lipoprotein lipase (LPL), cause a drastic reduction of serum lipoproteins and protect against the development of atherosclerotic cardiovascular disease. Therefore, ANGPTL3 is a promising therapy target. We characterized the impacts of ANGPTL3 depletion on the immortalized human hepatocyte (IHH) transcriptome, lipidome and human plasma lipoprotein lipidome. The transcriptome of ANGPTL3 knock-down (KD) cells showed altered expression of several pathways related to lipid metabolism. -

Glycerol Kinase Deficiency Alters Expression of Genes Involved in Lipid Metabolism, Carbohydrate Metabolism, and Insulin Signaling
European Journal of Human Genetics (2007) 15, 646–657 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg ARTICLE Glycerol kinase deficiency alters expression of genes involved in lipid metabolism, carbohydrate metabolism, and insulin signaling Lola Rahib1, Nicole K MacLennan2, Steve Horvath3,4, James C Liao5 and Katrina M Dipple*,1,2,4 1Biomedical Engineering, Interdepartmental Program, Henry Samueli School of Engineering and Applied Science at UCLA, Los Angeles, CA, USA; 2Department of Pediatrics and Mattel Children’s Hospital at UCLA, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA; 3Department of Biostatistics, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA; 4Department of Human Genetics, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA; 5Department of Chemical and Biomolecular Engineering, Henry Samueli School of Engineering and Applied Science at UCLA, Los Angeles, CA, USA Glycerol kinase (GK) is at the interface of fat and carbohydrate metabolism and has been implicated in insulin resistance and type 2 diabetes mellitus. To define GK’s role in insulin resistance, we examined gene expression in brown adipose tissue in a glycerol kinase knockout (KO) mouse model using microarray analysis. Global gene expression profiles of KO mice were distinct from wild type with 668 differentially expressed genes. These include genes involved in lipid metabolism, carbohydrate metabolism, insulin signaling, and insulin resistance. Real-time polymerase chain reaction analysis confirmed the differential expression of selected genes involved in lipid and carbohydrate metabolism. PathwayAssist analysis confirmed direct and indirect connections between glycerol kinase and genes in lipid metabolism, carbohydrate metabolism, insulin signaling, and insulin resistance.