Differences in Metabolomic and Transcriptomic Profiles Between
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

Implications in Parkinson's Disease
Journal of Clinical Medicine Review Lysosomal Ceramide Metabolism Disorders: Implications in Parkinson’s Disease Silvia Paciotti 1,2 , Elisabetta Albi 3 , Lucilla Parnetti 1 and Tommaso Beccari 3,* 1 Laboratory of Clinical Neurochemistry, Department of Medicine, University of Perugia, Sant’Andrea delle Fratte, 06132 Perugia, Italy; [email protected] (S.P.); [email protected] (L.P.) 2 Section of Physiology and Biochemistry, Department of Experimental Medicine, University of Perugia, Sant’Andrea delle Fratte, 06132 Perugia, Italy 3 Department of Pharmaceutical Sciences, University of Perugia, Via Fabretti, 06123 Perugia, Italy; [email protected] * Correspondence: [email protected] Received: 29 January 2020; Accepted: 20 February 2020; Published: 21 February 2020 Abstract: Ceramides are a family of bioactive lipids belonging to the class of sphingolipids. Sphingolipidoses are a group of inherited genetic diseases characterized by the unmetabolized sphingolipids and the consequent reduction of ceramide pool in lysosomes. Sphingolipidoses include several disorders as Sandhoff disease, Fabry disease, Gaucher disease, metachromatic leukodystrophy, Krabbe disease, Niemann Pick disease, Farber disease, and GM2 gangliosidosis. In sphingolipidosis, lysosomal lipid storage occurs in both the central nervous system and visceral tissues, and central nervous system pathology is a common hallmark for all of them. Parkinson’s disease, the most common neurodegenerative movement disorder, is characterized by the accumulation and aggregation of misfolded α-synuclein that seem associated to some lysosomal disorders, in particular Gaucher disease. This review provides evidence into the role of ceramide metabolism in the pathophysiology of lysosomes, highlighting the more recent findings on its involvement in Parkinson’s disease. Keywords: ceramide metabolism; Parkinson’s disease; α-synuclein; GBA; GLA; HEX A-B; GALC; ASAH1; SMPD1; ARSA * Correspondence [email protected] 1. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Silencing of Phosphoinositide-Specific
ANTICANCER RESEARCH 34: 4069-4076 (2014) Silencing of Phosphoinositide-specific Phospholipase C ε Remodulates the Expression of the Phosphoinositide Signal Transduction Pathway in Human Osteosarcoma Cell Lines VINCENZA RITA LO VASCO1, MARTINA LEOPIZZI2, DANIELA STOPPOLONI3 and CARLO DELLA ROCCA2 Departments of 1Sense Organs , 2Medicine and Surgery Sciences and Biotechnologies and 3Biochemistry Sciences “A. Rossi Fanelli”, Sapienza University, Rome, Italy Abstract. Background: Ezrin, a member of the signal transduction pathway (5). The reduction of PIP2 ezrin–radixin–moesin family, is involved in the metastatic induces ezrin dissociation from the plasma membrane (6). spread of osteosarcoma. Ezrin binds phosphatydil inositol-4,5- The levels of PIP2 are regulated by the PI-specific bisphosphate (PIP2), a crucial molecule of the phospholipase C (PI-PLC) family (7), constituting thirteen phosphoinositide signal transduction pathway. PIP2 levels are enzymes divided into six sub-families on the basis of amino regulated by phosphoinositide-specific phospholipase C (PI- acid sequence, domain structure, mechanism of recruitment PLC) enzymes. PI-PLCε isoform, a well-characterized direct and tissue distribution (7-15). PI-PLCε, a direct effector of effector of rat sarcoma (RAS), is at a unique convergence RAS (14-15), might be the point of convergence for the point for the broad range of signaling pathways that promote broad range of signalling pathways that promote the RAS GTPase-mediated signalling. Materials and Methods. By RASGTPase-mediated signalling (16). using molecular biology methods and microscopic analyses, In previous studies, we suggested a relationship between we analyzed the expression of ezrin and PLC genes after PI-PLC expression and ezrin (17-18). -

The Role of Phospholipases A2 in Schizophrenia
Molecular Psychiatry (2006) 11, 547–556 & 2006 Nature Publishing Group All rights reserved 1359-4184/06 $30.00 www.nature.com/mp FEATURE REVIEW The role of phospholipases A2 in schizophrenia MH Law1, RGH Cotton1 and GE Berger1,2 1Genomic Disorders Research Centre, Melbourne, VI, Australia and 2ORYGEN Research Centre, Melbourne, VI, Australia A range of neurotransmitter systems have been implicated in the pathogenesis of schizophrenia based on the antidopaminergic activities of antipsychotic medications, and chemicals that can induce psychotic-like symptoms, such as ketamine or PCP. Such neurotransmitter systems often mediate their cellular response via G-protein-coupled release of arachidonic acid (AA) via the activation of phospholipases A2 (PLA2s). The interaction of three PLA2s are important for the regulation of the release of AA – phospholipase A2 Group 2 A, phospholipase A2 Group 4A and phospholipase A2 Group 6A. Gene variations of these three key enzymes have been associated with schizophrenia with conflicting results. Preclinical data suggest that the activity of these three enzymes are associated with monoaminergic neurotransmission, and may contribute to the differential efficacy of antipsychotic medications, as well as other biological changes thought to underlie schizophrenia, such as altered neurodevelopment and synaptic remodelling. We review the evidence and discuss the potential roles of these three key enzymes for schizophrenia with particular emphasis on published association studies. Molecular Psychiatry (2006) 11, 547–556. doi:10.1038/sj.mp.4001819; published online 4 April 2006 Keywords: review; schizophrenia/genetics; PLA2/genetics; arachidonic acid; common diseases; PLA2/schizophrenia Introduction (PLA2GVIA, PLA2G6A), dopamine, serotonin, G- protein-coupled receptor, eicosanoids and phospho- Neurotransmitters, such as dopamine, serotonin and lipids. -

The Role of Genetic Variation in Predisposition to Alcohol-Related Chronic Pancreatitis
The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy by Marianne Lucy Johnstone April 2015 The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis 2015 Abstract Background Chronic pancreatitis (CP) is a disease of fibrosis of the pancreas for which alcohol is the main causative agent. However, only a small proportion of alcoholics develop chronic pancreatitis. Genetic polymorphism may affect pancreatitis risk. Aim To determine the factors required to classify a chronic pancreatic population and identify genetic variations that may explain why only some alcoholics develop chronic pancreatitis. Methods The most appropriate method of diagnosing CP was assessed using a systematic review. Genetics of different populations of alcohol-related chronic pancreatitics (ACP) were explored using four different techniques: genome-wide association study (GWAS); custom arrays; PCR of variable nucleotide tandem repeats (VNTR) and next generation sequencing (NGS) of selected genes. Results EUS and sMR were identified as giving the overall best sensitivity and specificity for diagnosing CP. GWAS revealed two associations with CP (identified and replicated) at PRSS1-PRSS2_rs10273639 (OR 0.73, 95% CI 0.68-0.79) and X-linked CLDN2_rs12688220 (OR 1.39, 1.28-1.49) and the association was more pronounced in the ACP group (OR 0.56, 0.48-0.64)and OR 2.11, 1.84-2.42). The previously identified VNTR in CEL was shown to have a lower frequency of the normal repeat in ACP than alcoholic liver disease (ALD; OR 0.61, 0.41-0.93). -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -
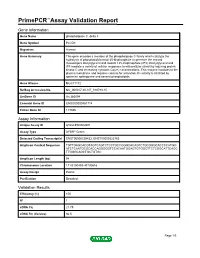
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name phospholipase C, delta 3 Gene Symbol PLCD3 Organism Human Gene Summary This gene encodes a member of the phospholipase C family which catalyze the hydrolysis of phosphatidylinositol 45-bisphosphate to generate the second messengers diacylglycerol and inositol 145-trisphosphate (IP3). Diacylglycerol and IP3 mediate a variety of cellular responses to extracellular stimuli by inducing protein kinase C and increasing cytosolic Ca(2+) concentrations. This enzyme localizes to the plasma membrane and requires calcium for activation. Its activity is inhibited by spermine sphingosine and several phospholipids. Gene Aliases MGC71172 RefSeq Accession No. NC_000017.10, NT_010783.15 UniGene ID Hs.380094 Ensembl Gene ID ENSG00000161714 Entrez Gene ID 113026 Assay Information Unique Assay ID qHsaCED0002601 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000539433, ENST00000322765 Amplicon Context Sequence TGTTGAGCACGTAGTCAGTCTCCTGCCGGGCACAGTCTGCGGGCACCCCATGG ATCTCAATGCGCACCAGGGGGTCCACAATGGAGTGTGGCTTCTCGGCATTCAGC TTGGGCAGCTGCTGTGC Amplicon Length (bp) 94 Chromosome Location 17:43190493-43190616 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 1 cDNA Cq 21.75 cDNA Tm (Celsius) 86.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 23.62 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PLCD3, Human Amplification Plot Amplification of cDNA generated from -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Genetic Variants in PLCB4/PLCB1 As Susceptibility Loci for Coronary Artery
www.nature.com/scientificreports OPEN Genetic variants in PLCB4/PLCB1 as susceptibility loci for coronary artery aneurysm formation in Received: 27 March 2015 Accepted: 04 September 2015 Kawasaki disease in Han Chinese Published: 05 October 2015 in Taiwan Ying-Ju Lin1,2, Jeng-Sheng Chang3,4, Xiang Liu5, Hsinyi Tsang5, Wen-Kuei Chien6, Jin-Hua Chen6,7, Hsin-Yang Hsieh3,8, Kai-Chung Hsueh9, Yi-Tzone Shiao10, Ju-Pi Li2,11, Cheng-Wen Lin12, Chih-Ho Lai13, Jer-Yuarn Wu2,14, Chien-Hsiun Chen2,14, Jaung-Geng Lin2, Ting-Hsu Lin1, Chiu-Chu Liao1, Shao-Mei Huang1, Yu-Ching Lan15, Tsung-Jung Ho2, Wen-Miin Liang16, Yi-Chun Yeh16, Jung-Chun Lin17 & Fuu-Jen Tsai1,2,18 Kawasaki disease (KD) is an acute, inflammatory, and self-limited vasculitis affecting infants and young children. Coronary artery aneurysm (CAA) formation is the major complication of KD and the leading cause of acquired cardiovascular disease among children. To identify susceptible loci that might predispose patients with KD to CAA formation, a genome-wide association screen was performed in a Taiwanese KD cohort. Patients with both KD and CAA had longer fever duration and delayed intravenous immunoglobulin treatment time. After adjusting for these factors, 100 susceptibility loci were identified. Four genes were identified from a single cluster of 35 using the Ingenuity Pathway Analysis (IPA) Knowledge Base. Silencing KCNQ5, PLCB1, PLCB4, and PLCL1 inhibited the effect of lipopolysaccharide-induced endothelial cell inflammation with varying degrees of proinflammatory cytokine expression. PLCB1 showed the most significant inhibition. Endothelial cell inflammation was also inhibited by using a phospholipase C (PLC) inhibitor.