Phospholipases of Mineralization Competent Cells and Matrix Vesicles: Roles in Physiological and Pathological Mineralizations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Figure 1. Vimentin
Double mutant specific genes Transcript gene_assignment Gene Symbol RefSeq FDR Fold- FDR Fold- FDR Fold- ID (single vs. Change (double Change (double Change wt) (single vs. wt) (double vs. single) (double vs. wt) vs. wt) vs. single) 10485013 BC085239 // 1110051M20Rik // RIKEN cDNA 1110051M20 gene // 2 E1 // 228356 /// NM 1110051M20Ri BC085239 0.164013 -1.38517 0.0345128 -2.24228 0.154535 -1.61877 k 10358717 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 /// BC 1700025G04Rik NM_197990 0.142593 -1.37878 0.0212926 -3.13385 0.093068 -2.27291 10358713 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 1700025G04Rik NM_197990 0.0655213 -1.71563 0.0222468 -2.32498 0.166843 -1.35517 10481312 NM_027283 // 1700026L06Rik // RIKEN cDNA 1700026L06 gene // 2 A3 // 69987 /// EN 1700026L06Rik NM_027283 0.0503754 -1.46385 0.0140999 -2.19537 0.0825609 -1.49972 10351465 BC150846 // 1700084C01Rik // RIKEN cDNA 1700084C01 gene // 1 H3 // 78465 /// NM_ 1700084C01Rik BC150846 0.107391 -1.5916 0.0385418 -2.05801 0.295457 -1.29305 10569654 AK007416 // 1810010D01Rik // RIKEN cDNA 1810010D01 gene // 7 F5 // 381935 /// XR 1810010D01Rik AK007416 0.145576 1.69432 0.0476957 2.51662 0.288571 1.48533 10508883 NM_001083916 // 1810019J16Rik // RIKEN cDNA 1810019J16 gene // 4 D2.3 // 69073 / 1810019J16Rik NM_001083916 0.0533206 1.57139 0.0145433 2.56417 0.0836674 1.63179 10585282 ENSMUST00000050829 // 2010007H06Rik // RIKEN cDNA 2010007H06 gene // --- // 6984 2010007H06Rik ENSMUST00000050829 0.129914 -1.71998 0.0434862 -2.51672 -

Genazzani-2003-Calcineurin And
Edinburgh Research Explorer Calcineurin controls the expression of numerous genes in cerebellar granule cells Citation for published version: Kramer, D, Fresu, L, Ashby, DS, Freeman, TC & Genazzani, AA 2003, 'Calcineurin controls the expression of numerous genes in cerebellar granule cells', Molecular and Cellular Neuroscience, vol. 23, no. 2, pp. 325- 30. Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Molecular and Cellular Neuroscience Publisher Rights Statement: © 2003 Elsevier Science (USA) General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 Molecular and Cellular Neuroscience 23 (2003) 325–330 www.elsevier.com/locate/ymcne Calcineurin controls the expression of numerous genes in cerebellar granule cells Dana Kramer,a Luigia Fresu,b Dominique S. Ashby,a Tom C. Freeman,c and Armando A. Genazzania,* a Department of Pharmacology, Tennis Court Road, Cambridge, CB2 1PD, UK b Department of Pharmacology, Gynaecology and Obstetrics, University of Sassari, Italy c Microarray Group, MRC HGMP-RC, Wellcome Trust Genome Campus, Hinxton, CB10 1SB, UK Received 31 October 2002; revised 7 January 2003; accepted 20 January 2003 Abstract The Ca2ϩ/calmodulin-dependent phosphatase calcineurin plays a crucial role in gene expression in different cell types such as T-lymphocytes, cardiac myocytes, and smooth muscle cells. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Silencing of Phosphoinositide-Specific
ANTICANCER RESEARCH 34: 4069-4076 (2014) Silencing of Phosphoinositide-specific Phospholipase C ε Remodulates the Expression of the Phosphoinositide Signal Transduction Pathway in Human Osteosarcoma Cell Lines VINCENZA RITA LO VASCO1, MARTINA LEOPIZZI2, DANIELA STOPPOLONI3 and CARLO DELLA ROCCA2 Departments of 1Sense Organs , 2Medicine and Surgery Sciences and Biotechnologies and 3Biochemistry Sciences “A. Rossi Fanelli”, Sapienza University, Rome, Italy Abstract. Background: Ezrin, a member of the signal transduction pathway (5). The reduction of PIP2 ezrin–radixin–moesin family, is involved in the metastatic induces ezrin dissociation from the plasma membrane (6). spread of osteosarcoma. Ezrin binds phosphatydil inositol-4,5- The levels of PIP2 are regulated by the PI-specific bisphosphate (PIP2), a crucial molecule of the phospholipase C (PI-PLC) family (7), constituting thirteen phosphoinositide signal transduction pathway. PIP2 levels are enzymes divided into six sub-families on the basis of amino regulated by phosphoinositide-specific phospholipase C (PI- acid sequence, domain structure, mechanism of recruitment PLC) enzymes. PI-PLCε isoform, a well-characterized direct and tissue distribution (7-15). PI-PLCε, a direct effector of effector of rat sarcoma (RAS), is at a unique convergence RAS (14-15), might be the point of convergence for the point for the broad range of signaling pathways that promote broad range of signalling pathways that promote the RAS GTPase-mediated signalling. Materials and Methods. By RASGTPase-mediated signalling (16). using molecular biology methods and microscopic analyses, In previous studies, we suggested a relationship between we analyzed the expression of ezrin and PLC genes after PI-PLC expression and ezrin (17-18). -

Gentaur Products List
Chapter 2 : Gentaur Products List • Rabbit Anti LAMR1 Polyclonal Antibody Cy5 Conjugated Conjugated • Rabbit Anti Podoplanin gp36 Polyclonal Antibody Cy5 • Rabbit Anti LAMR1 CT Polyclonal Antibody Cy5 • Rabbit Anti phospho NFKB p65 Ser536 Polyclonal Conjugated Conjugated Antibody Cy5 Conjugated • Rabbit Anti CHRNA7 Polyclonal Antibody Cy5 Conjugated • Rat Anti IAA Monoclonal Antibody Cy5 Conjugated • Rabbit Anti EV71 VP1 CT Polyclonal Antibody Cy5 • Rabbit Anti Connexin 40 Polyclonal Antibody Cy5 • Rabbit Anti IAA Indole 3 Acetic Acid Polyclonal Antibody Conjugated Conjugated Cy5 Conjugated • Rabbit Anti LHR CGR Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Integrin beta 7 Polyclonal Antibody Cy5 • Rabbit Anti Natrexone Polyclonal Antibody Cy5 Conjugated • Rabbit Anti MMP 20 Polyclonal Antibody Cy5 Conjugated Conjugated • Rabbit Anti Melamine Polyclonal Antibody Cy5 Conjugated • Rabbit Anti BCHE NT Polyclonal Antibody Cy5 Conjugated • Rabbit Anti NAP1 NAP1L1 Polyclonal Antibody Cy5 • Rabbit Anti Acetyl p53 K382 Polyclonal Antibody Cy5 • Rabbit Anti BCHE CT Polyclonal Antibody Cy5 Conjugated Conjugated Conjugated • Rabbit Anti HPV16 E6 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti CCP Polyclonal Antibody Cy5 Conjugated • Rabbit Anti JAK2 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti HPV18 E6 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti HDC Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Microsporidia protien Polyclonal Antibody Cy5 • Rabbit Anti HPV16 E7 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Neurocan Polyclonal -

The Role of Genetic Variation in Predisposition to Alcohol-Related Chronic Pancreatitis
The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy by Marianne Lucy Johnstone April 2015 The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis 2015 Abstract Background Chronic pancreatitis (CP) is a disease of fibrosis of the pancreas for which alcohol is the main causative agent. However, only a small proportion of alcoholics develop chronic pancreatitis. Genetic polymorphism may affect pancreatitis risk. Aim To determine the factors required to classify a chronic pancreatic population and identify genetic variations that may explain why only some alcoholics develop chronic pancreatitis. Methods The most appropriate method of diagnosing CP was assessed using a systematic review. Genetics of different populations of alcohol-related chronic pancreatitics (ACP) were explored using four different techniques: genome-wide association study (GWAS); custom arrays; PCR of variable nucleotide tandem repeats (VNTR) and next generation sequencing (NGS) of selected genes. Results EUS and sMR were identified as giving the overall best sensitivity and specificity for diagnosing CP. GWAS revealed two associations with CP (identified and replicated) at PRSS1-PRSS2_rs10273639 (OR 0.73, 95% CI 0.68-0.79) and X-linked CLDN2_rs12688220 (OR 1.39, 1.28-1.49) and the association was more pronounced in the ACP group (OR 0.56, 0.48-0.64)and OR 2.11, 1.84-2.42). The previously identified VNTR in CEL was shown to have a lower frequency of the normal repeat in ACP than alcoholic liver disease (ALD; OR 0.61, 0.41-0.93). -

Survival-Associated Metabolic Genes in Colon and Rectal Cancers
Survival-associated Metabolic Genes in Colon and Rectal Cancers Yanfen Cui ( [email protected] ) Tianjin Cancer Institute: Tianjin Tumor Hospital https://orcid.org/0000-0001-7760-7503 Baoai Han tianjin tumor hospital He Zhang tianjin tumor hospital Zhiyong Wang tianjin tumor hospital Hui Liu tianjin tumor hospital Fei Zhang tianjin tumor hospital Ruifang Niu tianjin tumor hospital Research Keywords: colon cancer, rectal cancer, prognosis, metabolism Posted Date: December 4th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-117478/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/42 Abstract Background Uncontrolled proliferation is the most prominent biological feature of tumors. To rapidly proliferate and maximize the use of available nutrients, tumor cells regulate their metabolic behavior and the expression of metabolism-related genes (MRGs). In this study, we aimed to construct prognosis models for colon and rectal cancers, using MRGs to indicate the prognoses of patients. Methods We rst acquired the gene expression proles of colon and rectal cancers from the TCGA and GEO database, and utilized univariate Cox analysis, lasso regression, and multivariable cox analysis to identify MRGs for risk models. Then GSEA and KEGG functional enrichment analysis were utilized to identify the metabolism pathway of MRGs in the risk models and analyzed these genes comprehensively using GSCALite. Results Eight genes (CPT1C, PLCB2, PLA2G2D, GAMT, ENPP2, PIP4K2B, GPX3, and GSR) in the colon cancer risk model and six genes (TDO2, PKLR, GAMT, EARS2, ACO1, and WAS) in the rectal cancer risk model were identied successfully. Multivariate Cox analysis indicated that the models predicted overall survival accurately and independently for patients with colon or rectal cancer. -

Effective Angiogenesis Requires Regulation of Phosphoinositide Signaling
Effective angiogenesis requires regulation of phosphoinositide signaling Elizabeth Michele Davies, Rajendra Gurung, Kai Qin Le, Christina Anne Mitchell* Cancer Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Victoria 3800, Australia *Corresponding author [email protected] Abstract Phosphoinositide signaling regulates numerous downstream effectors that mediate cellular processes which influence cell cycle progression, migration, proliferation, growth, survival, metabolism and vesicular trafficking. A prominent role for phosphoinositide 3-kinase, which generates phosphatidylinositol 3,4,5-trisphosphate, a phospholipid that activates a plethora of effectors including AKT and FOXO during embryonic and postnatal angiogenesis, has been described. In addition, phosphatidylinositol 3-phosphate signaling is required for endosomal trafficking, which contributes to vascular remodeling. This review will examine the role phosphoinositide signaling plays in the endothelium and its contribution to sprouting angiogenesis. Keywords Angiogenesis, Phosphoinositide, PI3K, PTEN 1. Introduction Establishment of a functional vasculature is essential for embryonic development, and also for the maintenance of tissue regeneration, inflammatory responses and wound healing in the adult. Vascular development is characterized by two distinct physiological processes, vasculogenesis and angiogenesis. Vasculogenesis is predominantly limited to embryonic development, whereby differentiation -
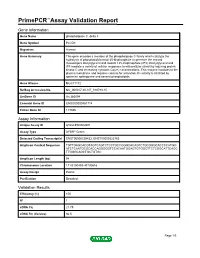
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name phospholipase C, delta 3 Gene Symbol PLCD3 Organism Human Gene Summary This gene encodes a member of the phospholipase C family which catalyze the hydrolysis of phosphatidylinositol 45-bisphosphate to generate the second messengers diacylglycerol and inositol 145-trisphosphate (IP3). Diacylglycerol and IP3 mediate a variety of cellular responses to extracellular stimuli by inducing protein kinase C and increasing cytosolic Ca(2+) concentrations. This enzyme localizes to the plasma membrane and requires calcium for activation. Its activity is inhibited by spermine sphingosine and several phospholipids. Gene Aliases MGC71172 RefSeq Accession No. NC_000017.10, NT_010783.15 UniGene ID Hs.380094 Ensembl Gene ID ENSG00000161714 Entrez Gene ID 113026 Assay Information Unique Assay ID qHsaCED0002601 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000539433, ENST00000322765 Amplicon Context Sequence TGTTGAGCACGTAGTCAGTCTCCTGCCGGGCACAGTCTGCGGGCACCCCATGG ATCTCAATGCGCACCAGGGGGTCCACAATGGAGTGTGGCTTCTCGGCATTCAGC TTGGGCAGCTGCTGTGC Amplicon Length (bp) 94 Chromosome Location 17:43190493-43190616 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 1 cDNA Cq 21.75 cDNA Tm (Celsius) 86.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 23.62 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PLCD3, Human Amplification Plot Amplification of cDNA generated from -

Supplementary Table 1 List of 335 Genes Differentially Expressed Between Primary (P) and Metastatic (M) Tumours
Supplementary Table 1 List of 335 genes differentially expressed between primary (P) and metastatic (M) tumours Spot ID I.M.A.G.E. UniGene Symbol Name Clone ID Cluster 296529 296529 In multiple clusters 731356 731356 Hs.140452 M6PRBP1 mannose-6-phosphate receptor binding protein 1 840942 840942 Hs.368409 HLA-DPB1 major histocompatibility complex, class II, DP beta 1 142122 142122 Hs.115912 AFAP actin filament associated protein 1891918 1891918 Hs.90073 CSE1L CSE1 chromosome segregation 1-like (yeast) 1323432 1323432 Hs.303154 IDS iduronate 2-sulfatase (Hunter syndrome) 788566 788566 Hs.80296 PCP4 Purkinje cell protein 4 591281 591281 Hs.80680 MVP major vault protein 815530 815530 Hs.172813 ARHGEF7 Rho guanine nucleotide exchange factor (GEF) 7 825312 825312 Hs.246310 ATP5J ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 784830 784830 Hs.412842 C10orf7 chromosome 10 open reading frame 7 840878 840878 Hs.75616 DHCR24 24-dehydrocholesterol reductase 669443 669443 Hs.158195 HSF2 heat shock transcription factor 2 2485436 2485436 Data not found 82903 82903 Hs.370937 TAPBP TAP binding protein (tapasin) 771258 771258 Hs.85258 CD8A CD8 antigen, alpha polypeptide (p32) 85128 85128 Hs.8986 C1QB complement component 1, q subcomponent, beta polypeptide 41929 41929 Hs.39252 PICALM phosphatidylinositol binding clathrin assembly protein 148469 148469 Hs.9963 TYROBP TYRO protein tyrosine kinase binding protein 415145 415145 Hs.1376 HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 810017 810017 Hs.179657 PLAUR plasminogen activator, -

Genetic Defect in Phospholipase Cd1 Protects Mice from Obesity By
ORIGINAL ARTICLE Genetic Defect in Phospholipase Cd1 Protects Mice From Obesity by Regulating Thermogenesis and Adipogenesis Masayuki Hirata,1 Mutsumi Suzuki,1 Rika Ishii,1 Reiko Satow,1 Takafumi Uchida,1 Tomoya Kitazumi,2 Tsutomu Sasaki,2 Tadahiro Kitamura,2 Hideki Yamaguchi,3,4 Yoshikazu Nakamura,1 and Kiyoko Fukami1 OBJECTIVE—Regulation of obesity development is an impor- size of adipocytes is a hallmark of obesity. The former tant issue to prevent metabolic syndromes. Gene-disrupted mice seems to be caused by proliferation and differentiation of of phospholipase Cd1(PLCd1), a key enzyme of phosphoinositide preadipocytes. On the other hand, the diet-induced in- turnover, seemed to show leanness. Here we examined whether crease in cell size is characterized by adipocyte hypertro- and how PLCd1 is involved in obesity. phy, which may be primarily caused by excessive lipid RESEARCH DESIGN AND METHODS—Weight gain, insulin overload and a decrease in metabolic rate. sensitivity, and metabolic rate in PLCd12/2 mice were compared Brown adipose tissue (BAT) is implicated in thermo- with PLCd1+/2 littermate mice on a high-fat diet. Thermogenic genesis and metabolic enhancement (5). Recent reports in- and adipogenetic potentials of PLCd12/2 immortalized brown dicated that BAT and skeletal muscle originate from adipocytes and adipogenesis of PLCd1-knockdown (KD) 3T3L1 a common precursor cell (6–9). Like skeletal muscle, BAT 2 2 cells, or PLCd1 / white adipose tissue (WAT) stromal-vascular plays a role in thermogenesis by promoting the expression of fraction (SVF) cells, were also investigated. a thermogenic gene, uncoupling protein 1 (UCP1). Upregu- RESULTS—PLCd12/2 mice showed marked decreases in weight lation of UCP1 by genetic manipulations or pharmacological gain and mass of epididymal WAT and preserved insulin sensitivity agents has been shown to reduce obesity and improve in- compared with PLCd1+/2 mice on a high-fat diet. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase