Downloads/Manuals/Wt Sensetarget Label Manual.Pdf)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Silencing of Phosphoinositide-Specific
ANTICANCER RESEARCH 34: 4069-4076 (2014) Silencing of Phosphoinositide-specific Phospholipase C ε Remodulates the Expression of the Phosphoinositide Signal Transduction Pathway in Human Osteosarcoma Cell Lines VINCENZA RITA LO VASCO1, MARTINA LEOPIZZI2, DANIELA STOPPOLONI3 and CARLO DELLA ROCCA2 Departments of 1Sense Organs , 2Medicine and Surgery Sciences and Biotechnologies and 3Biochemistry Sciences “A. Rossi Fanelli”, Sapienza University, Rome, Italy Abstract. Background: Ezrin, a member of the signal transduction pathway (5). The reduction of PIP2 ezrin–radixin–moesin family, is involved in the metastatic induces ezrin dissociation from the plasma membrane (6). spread of osteosarcoma. Ezrin binds phosphatydil inositol-4,5- The levels of PIP2 are regulated by the PI-specific bisphosphate (PIP2), a crucial molecule of the phospholipase C (PI-PLC) family (7), constituting thirteen phosphoinositide signal transduction pathway. PIP2 levels are enzymes divided into six sub-families on the basis of amino regulated by phosphoinositide-specific phospholipase C (PI- acid sequence, domain structure, mechanism of recruitment PLC) enzymes. PI-PLCε isoform, a well-characterized direct and tissue distribution (7-15). PI-PLCε, a direct effector of effector of rat sarcoma (RAS), is at a unique convergence RAS (14-15), might be the point of convergence for the point for the broad range of signaling pathways that promote broad range of signalling pathways that promote the RAS GTPase-mediated signalling. Materials and Methods. By RASGTPase-mediated signalling (16). using molecular biology methods and microscopic analyses, In previous studies, we suggested a relationship between we analyzed the expression of ezrin and PLC genes after PI-PLC expression and ezrin (17-18). -

Effective Angiogenesis Requires Regulation of Phosphoinositide Signaling
Effective angiogenesis requires regulation of phosphoinositide signaling Elizabeth Michele Davies, Rajendra Gurung, Kai Qin Le, Christina Anne Mitchell* Cancer Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Victoria 3800, Australia *Corresponding author [email protected] Abstract Phosphoinositide signaling regulates numerous downstream effectors that mediate cellular processes which influence cell cycle progression, migration, proliferation, growth, survival, metabolism and vesicular trafficking. A prominent role for phosphoinositide 3-kinase, which generates phosphatidylinositol 3,4,5-trisphosphate, a phospholipid that activates a plethora of effectors including AKT and FOXO during embryonic and postnatal angiogenesis, has been described. In addition, phosphatidylinositol 3-phosphate signaling is required for endosomal trafficking, which contributes to vascular remodeling. This review will examine the role phosphoinositide signaling plays in the endothelium and its contribution to sprouting angiogenesis. Keywords Angiogenesis, Phosphoinositide, PI3K, PTEN 1. Introduction Establishment of a functional vasculature is essential for embryonic development, and also for the maintenance of tissue regeneration, inflammatory responses and wound healing in the adult. Vascular development is characterized by two distinct physiological processes, vasculogenesis and angiogenesis. Vasculogenesis is predominantly limited to embryonic development, whereby differentiation -
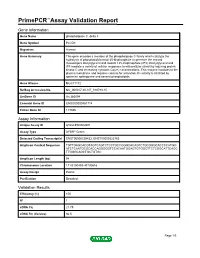
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name phospholipase C, delta 3 Gene Symbol PLCD3 Organism Human Gene Summary This gene encodes a member of the phospholipase C family which catalyze the hydrolysis of phosphatidylinositol 45-bisphosphate to generate the second messengers diacylglycerol and inositol 145-trisphosphate (IP3). Diacylglycerol and IP3 mediate a variety of cellular responses to extracellular stimuli by inducing protein kinase C and increasing cytosolic Ca(2+) concentrations. This enzyme localizes to the plasma membrane and requires calcium for activation. Its activity is inhibited by spermine sphingosine and several phospholipids. Gene Aliases MGC71172 RefSeq Accession No. NC_000017.10, NT_010783.15 UniGene ID Hs.380094 Ensembl Gene ID ENSG00000161714 Entrez Gene ID 113026 Assay Information Unique Assay ID qHsaCED0002601 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000539433, ENST00000322765 Amplicon Context Sequence TGTTGAGCACGTAGTCAGTCTCCTGCCGGGCACAGTCTGCGGGCACCCCATGG ATCTCAATGCGCACCAGGGGGTCCACAATGGAGTGTGGCTTCTCGGCATTCAGC TTGGGCAGCTGCTGTGC Amplicon Length (bp) 94 Chromosome Location 17:43190493-43190616 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 1 cDNA Cq 21.75 cDNA Tm (Celsius) 86.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 23.62 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PLCD3, Human Amplification Plot Amplification of cDNA generated from -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

Transcriptomic Uniqueness and Commonality of the Ion Channels and Transporters in the Four Heart Chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A
www.nature.com/scientificreports OPEN Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A. Iacobas3,4* Myocardium transcriptomes of left and right atria and ventricles from four adult male C57Bl/6j mice were profled with Agilent microarrays to identify the diferences responsible for the distinct functional roles of the four heart chambers. Female mice were not investigated owing to their transcriptome dependence on the estrous cycle phase. Out of the quantifed 16,886 unigenes, 15.76% on the left side and 16.5% on the right side exhibited diferential expression between the atrium and the ventricle, while 5.8% of genes were diferently expressed between the two atria and only 1.2% between the two ventricles. The study revealed also chamber diferences in gene expression control and coordination. We analyzed ion channels and transporters, and genes within the cardiac muscle contraction, oxidative phosphorylation, glycolysis/gluconeogenesis, calcium and adrenergic signaling pathways. Interestingly, while expression of Ank2 oscillates in phase with all 27 quantifed binding partners in the left ventricle, the percentage of in-phase oscillating partners of Ank2 is 15% and 37% in the left and right atria and 74% in the right ventricle. The analysis indicated high interventricular synchrony of the ion channels expressions and the substantially lower synchrony between the two atria and between the atrium and the ventricle from the same side. Starting with crocodilians, the heart pumps the blood through the pulmonary circulation and the systemic cir- culation by the coordinated rhythmic contractions of its upper lef and right atria (LA, RA) and lower lef and right ventricles (LV, RV). -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Novel Associations of Nonstructural Loci with Paraoxonase Activity
Hindawi Publishing Corporation Journal of Lipids Volume 2012, Article ID 189681, 7 pages doi:10.1155/2012/189681 Research Article Novel Associations of Nonstructural Loci with Paraoxonase Activity Ellen E. Quillen, David L. Rainwater, Thomas D. Dyer, Melanie A. Carless, Joanne E. Curran, Matthew P.Johnson, Harald H. H. Goring,¨ Shelley A. Cole, Sue Rutherford, Jean W.MacCluer, Eric K. Moses, John Blangero, Laura Almasy, and Michael C. Mahaney Department of Genetics, Texas Biomedical Research Institute, P.O. Box 760549, San Antonio, TX 78245-0549, USA Correspondence should be addressed to Ellen E. Quillen, [email protected] Received 22 January 2012; Accepted 19 February 2012 Academic Editor: Mira Rosenblat Copyright © 2012 Ellen E. Quillen et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The high-density-lipoprotein-(HDL-) associated esterase paraoxonase 1 (PON1) is a likely contributor to the antioxidant and antiatherosclerotic capabilities of HDL. Two nonsynonymous mutations in the structural gene, PON1, have been associated with variation in activity levels, but substantial interindividual differences remain unexplained and are greatest for substrates other than the eponymous paraoxon. PON1 activity levels were measured for three substrates—organophosphate paraoxon, arylester phenyl acetate, and lactone dihydrocoumarin—in 767 Mexican American individuals from San Antonio, Texas. Genetic influences on activity levels for each substrate were evaluated by association with approximately one million single nucleotide polymorphism (SNPs) while conditioning on PON1 genotypes. Significant associations were detected at five loci including regions on chromosomes 4 and 17 known to be associated with atherosclerosis and lipoprotein regulation and loci on chromosome 3 that regulate ubiquitous transcription factors. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

Changes in Gene Expression Under MET-Stress in CAPAN1 and CFPAC1
Supplement: Changes in gene expression under MET-stress in CAPAN1 and CFPAC1. Significant (<0.05) changes of 2.5 fold or greater that occur in cultures under MET-stress for either CAPAN1 or CFPAC for 4 and 6 d respectively, are listed. NE: not expressed denotes relative expression of less than 200 in both methionine (M) and homocysteine (H) sets. CAPAN1 CAPAN1 CFPAC1 CFPAC1 GENE SYMBOL CHROMOSOME MET HCYS M/H MET HCYS M/H Growth arrest and DNA-damage-inducible, beta GADD45B chr19p13.3 47.4 2605.4 0.02 546.5 978.9 0.56 clusterin CLU chr8p21-p12 31 610.2 0.05 NE cyclin-dependent kinase 5 CDK5 chr7q36 44 502.3 0.09 286.1 628.4 0.46 SMAD, mothers against DPP homolog 7 SMAD7 chr18q21.1 30.1 337.2 0.09 410.8 968.3 0.63 colony stimulating factor 1 (macrophage) CSF1 chr1p21-p13 50.4 487.1 0.10 1105.5 1741.8 0.42 breast cancer 1, early onset BRCA1 chr17q21 31.6 316.5 0.11 125.5 427.1 0.29 TGF, beta receptor associated protein 1 TGFBRAP1 chr2q12.1 72.9 648.9 0.11 NE tuftelin interacting protein 11 TFIP11 chr22q12.1 63.8 561.5 0.11 212.2 382 0.56 three prime repair exonuclease 1 TREX1 chr3p21.3-p21.2 39.8 310.1 0.13 59.5 310.8 0.19 v-ets erythroblastosis virus E26 oncogene homolog2 ETS2 chr21q22.3|21q22.2 105 807.1 0.13 615.9 414.2 1.49 growth differentiation factor 15 GDF15 chr19p13.1-13.2 275 1940.1 0.14 4961.8 6636.5 0.75 c-myc binding protein MYCBP chr1p33-p32.2 32.4 226.8 0.14 448.6 525.5 0.85 protein (peptidyl-prolyl cis/trans isomerase) 1 PIN1 chr19p13 102 683.8 0.15 NE putative lymphocyte G0/G1 switch gene G0S2 chr1q32.2-q41 1485.3 9136.8 -

Potent Lipolytic Activity of Lactoferrin in Mature Adipocytes
Biosci. Biotechnol. Biochem., 77 (3), 566–571, 2013 Potent Lipolytic Activity of Lactoferrin in Mature Adipocytes y Tomoji ONO,1;2; Chikako FUJISAKI,1 Yasuharu ISHIHARA,1 Keiko IKOMA,1;2 Satoru MORISHITA,1;3 Michiaki MURAKOSHI,1;4 Keikichi SUGIYAMA,1;5 Hisanori KATO,3 Kazuo MIYASHITA,6 Toshihide YOSHIDA,4;7 and Hoyoku NISHINO4;5 1Research and Development Headquarters, Lion Corporation, 100 Tajima, Odawara, Kanagawa 256-0811, Japan 2Department of Supramolecular Biology, Graduate School of Nanobioscience, Yokohama City University, 3-9 Fukuura, Kanazawa-ku, Yokohama, Kanagawa 236-0004, Japan 3Food for Life, Organization for Interdisciplinary Research Projects, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan 4Kyoto Prefectural University of Medicine, Kawaramachi-Hirokoji, Kamigyou-ku, Kyoto 602-8566, Japan 5Research Organization of Science and Engineering, Ritsumeikan University, 1-1-1 Nojihigashi, Kusatsu, Shiga 525-8577, Japan 6Department of Marine Bioresources Chemistry, Faculty of Fisheries Sciences, Hokkaido University, 3-1-1 Minatocho, Hakodate, Hokkaido 041-8611, Japan 7Kyoto City Hospital, 1-2 Higashi-takada-cho, Mibu, Nakagyou-ku, Kyoto 604-8845, Japan Received October 22, 2012; Accepted November 26, 2012; Online Publication, March 7, 2013 [doi:10.1271/bbb.120817] Lactoferrin (LF) is a multifunctional glycoprotein resistance, high blood pressure, and dyslipidemia. To found in mammalian milk. We have shown in a previous prevent progression of metabolic syndrome, lifestyle clinical study that enteric-coated bovine LF tablets habits must be improved to achieve a balance between decreased visceral fat accumulation. To address the energy intake and consumption. In addition, the use of underlying mechanism, we conducted in vitro studies specific food factors as helpful supplements is attracting and revealed the anti-adipogenic action of LF in pre- increasing attention. -

Small Molecule Protein–Protein Interaction Inhibitors As CNS Therapeutic Agents: Current Progress and Future Hurdles
Neuropsychopharmacology REVIEWS (2009) 34, 126–141 & 2009 Nature Publishing Group All rights reserved 0893-133X/09 $30.00 ............................................................................................................................................................... REVIEW 126 www.neuropsychopharmacology.org Small Molecule Protein–Protein Interaction Inhibitors as CNS Therapeutic Agents: Current Progress and Future Hurdles 1 ,1,2,3 Levi L Blazer and Richard R Neubig* 1Department of Pharmacology, University of Michigan Medical School, Ann Arbor, MI, USA; 2Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI, USA; 3Center for Chemical Genomics, University of Michigan Medical School, Ann Arbor, MI, USA Protein–protein interactions are a crucial element in cellular function. The wealth of information currently available on intracellular-signaling pathways has led many to appreciate the untapped pool of potential drug targets that reside downstream of the commonly targeted receptors. Over the last two decades, there has been significant interest in developing therapeutics and chemical probes that inhibit specific protein–protein interactions. Although it has been a challenge to develop small molecules that are capable of occluding the large, often relatively featureless protein–protein interaction interface, there are increasing numbers of examples of small molecules that function in this manner with reasonable potency. This article will highlight the current progress in the development of small