Mullergliaregnerationtranscriptome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacillus Anthracis Edema Factor Substrate Specificity: Evidence for New Modes of Action
Toxins 2012, 4, 505-535; doi:10.3390/toxins4070505 OPEN ACCESS toxins ISSN 2072–6651 www.mdpi.com/journal/toxins Review Bacillus anthracis Edema Factor Substrate Specificity: Evidence for New Modes of Action Martin Göttle 1,*, Stefan Dove 2 and Roland Seifert 3 1 Department of Neurology, Emory University School of Medicine, 6302 Woodruff Memorial Research Building, 101 Woodruff Circle, Atlanta, GA 30322, USA 2 Department of Medicinal/Pharmaceutical Chemistry II, University of Regensburg, D-93040 Regensburg, Germany; E-Mail: [email protected] 3 Institute of Pharmacology, Medical School of Hannover, Carl-Neuberg-Str. 1, D-30625 Hannover, Germany; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-404-727-1678; Fax: +1-404-727-3157. Received: 23 April 2012; in revised form: 15 June 2012 / Accepted: 27 June 2012 / Published: 6 July 2012 Abstract: Since the isolation of Bacillus anthracis exotoxins in the 1960s, the detrimental activity of edema factor (EF) was considered as adenylyl cyclase activity only. Yet the catalytic site of EF was recently shown to accomplish cyclization of cytidine 5′-triphosphate, uridine 5′-triphosphate and inosine 5′-triphosphate, in addition to adenosine 5′-triphosphate. This review discusses the broad EF substrate specificity and possible implications of intracellular accumulation of cyclic cytidine 3′:5′-monophosphate, cyclic uridine 3′:5′-monophosphate and cyclic inosine 3′:5′-monophosphate on cellular functions vital for host defense. In particular, cAMP-independent mechanisms of action of EF on host cell signaling via protein kinase A, protein kinase G, phosphodiesterases and CNG channels are discussed. -

The Genome of Nanoarchaeum Equitans: Insights Into Early Archaeal Evolution and Derived Parasitism
The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism Elizabeth Waters†‡, Michael J. Hohn§, Ivan Ahel¶, David E. Graham††, Mark D. Adams‡‡, Mary Barnstead‡‡, Karen Y. Beeson‡‡, Lisa Bibbs†, Randall Bolanos‡‡, Martin Keller†, Keith Kretz†, Xiaoying Lin‡‡, Eric Mathur†, Jingwei Ni‡‡, Mircea Podar†, Toby Richardson†, Granger G. Sutton‡‡, Melvin Simon†, Dieter So¨ ll¶§§¶¶, Karl O. Stetter†§¶¶, Jay M. Short†, and Michiel Noordewier†¶¶ †Diversa Corporation, 4955 Directors Place, San Diego, CA 92121; ‡Department of Biology, San Diego State University, 5500 Campanile Drive, San Diego, CA 92182; §Lehrstuhl fu¨r Mikrobiologie und Archaeenzentrum, Universita¨t Regensburg, Universita¨tsstrasse 31, D-93053 Regensburg, Germany; ‡‡Celera Genomics Rockville, 45 West Gude Drive, Rockville, MD 20850; Departments of ¶Molecular Biophysics and Biochemistry and §§Chemistry, Yale University, New Haven, CT 06520-8114; and ʈDepartment of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061 Communicated by Carl R. Woese, University of Illinois at Urbana–Champaign, Urbana, IL, August 21, 2003 (received for review July 22, 2003) The hyperthermophile Nanoarchaeum equitans is an obligate sym- (6–8). Genomic DNA was either digested with restriction en- biont growing in coculture with the crenarchaeon Ignicoccus. zymes or sheared to provide clonable fragments. Two plasmid Ribosomal protein and rRNA-based phylogenies place its branching libraries were made by subcloning randomly sheared fragments point early in the archaeal lineage, representing the new archaeal of this DNA into a high-copy number vector (Ϸ2.8 kbp library) kingdom Nanoarchaeota. The N. equitans genome (490,885 base or low-copy number vector (Ϸ6.3 kbp library). DNA sequence pairs) encodes the machinery for information processing and was obtained from both ends of plasmid inserts to create repair, but lacks genes for lipid, cofactor, amino acid, or nucleotide ‘‘mate-pairs,’’ pairs of reads from single clones that should be biosyntheses. -

Genetic Expression Profile Analysis of the Temporal Inhibition of Quercetin and Naringenin on Lactobacillus Rhamnosus GG
robioti f P cs o & l a H n e r a u l t o h J Liu, et al., J Prob Health 2016, 4:2 Journal of Probiotics & Health DOI: 10.4172/2329-8901.1000139 ISSN: 2329-8901 Research Article Open Access Genetic Expression Profile Analysis of the Temporal Inhibition of Quercetin and Naringenin on Lactobacillus Rhamnosus GG Linshu Liu1*, Jenni Firrman1, Gustavo Arango Argoty2, Peggy Tomasula1, Masuko Kobori3, Liqing Zhang2 and Weidong Xiao4* 1Dairy and Functional Foods Research Unit, Eastern Regional Research Center, Agricultural Research Service, US Department of Agriculture, 600 E Mermaid Lane, Wyndmoor, PA 19038, USA 2Virginia Tech College of Engineering, Department of Computer Science, 1425 S Main St. Blacksburg, VA 24061, USA 3National Food Research Institute, National Agriculture and Food Research Organization, Tsukuba, Ibaraki 305-8642, Japan 4*Department of Microbiology and Immunology, Temple University School of Medicine, 3400 North Broad Street, Philadelphia, USA *Corresponding author: Weidong Xiao, Department of Microbiology and Immunology, Temple University School of Medicine, 3400 North Broad Street, Philadelphia, USA, Tel: 215-707-6392; E-mail: [email protected], LinShu Liu, Dairy and Functional Foods Research Unit, Eastern Regional Research Center, Agricultural Research Service, US Department of Agriculture, 600 E Mermaid Lane, Wyndmoor, PA 19038, USA. E-mail: [email protected] Received date: Jan 29, 2015; Accepted date: Feb 15, 2016; Published date: Feb 22, 2016 Copyright: © 2016 Liu LS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -
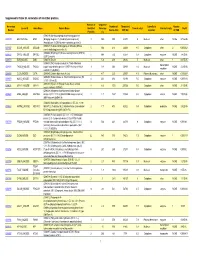
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase -

Dependent Protein Kinase to Cyclic CMP Agarose
Binding of Regulatory Subunits of Cyclic AMP- Dependent Protein Kinase to Cyclic CMP Agarose Andreas Hammerschmidt1., Bijon Chatterji2., Johannes Zeiser2, Anke Schro¨ der2, Hans- Gottfried Genieser3, Andreas Pich2, Volkhard Kaever1, Frank Schwede3, Sabine Wolter1", Roland Seifert1*" 1 Institute of Pharmacology, Hannover Medical School, Hannover, Germany, 2 Institute of Toxicology, Hannover Medical School, Hannover, Germany, 3 Biolog Life Science Institute, Bremen, Germany Abstract The bacterial adenylyl cyclase toxins CyaA from Bordetella pertussis and edema factor from Bacillus anthracis as well as soluble guanylyl cyclase a1b1 synthesize the cyclic pyrimidine nucleotide cCMP. These data raise the question to which effector proteins cCMP binds. Recently, we reported that cCMP activates the regulatory subunits RIa and RIIa of cAMP- dependent protein kinase. In this study, we used two cCMP agarose matrices as novel tools in combination with immunoblotting and mass spectrometry to identify cCMP-binding proteins. In agreement with our functional data, RIa and RIIa were identified as cCMP-binding proteins. These data corroborate the notion that cAMP-dependent protein kinase may serve as a cCMP target. Citation: Hammerschmidt A, Chatterji B, Zeiser J, Schro¨der A, Genieser H-G, et al. (2012) Binding of Regulatory Subunits of Cyclic AMP-Dependent Protein Kinase to Cyclic CMP Agarose. PLoS ONE 7(7): e39848. doi:10.1371/journal.pone.0039848 Editor: Andreas Hofmann, Griffith University, Australia Received May 11, 2012; Accepted May 31, 2012; Published July 9, 2012 Copyright: ß 2012 Hammerschmidt et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

1/05661 1 Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date _ . ... - 12 May 2011 (12.05.2011) W 2 11/05661 1 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12Q 1/00 (2006.0 1) C12Q 1/48 (2006.0 1) kind of national protection available): AE, AG, AL, AM, C12Q 1/42 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) Number: International Application DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/US20 10/054171 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 26 October 2010 (26.10.2010) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, (25) Filing Language: English SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, (26) Publication Language: English TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/255,068 26 October 2009 (26.10.2009) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, (71) Applicant (for all designated States except US): ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, MYREXIS, INC. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

AUSTRALIAN PATENT OFFICE (11) Application No. AU 199875933 B2
(12) PATENT (11) Application No. AU 199875933 B2 (19) AUSTRALIAN PATENT OFFICE (10) Patent No. 742342 (54) Title Nucleic acid arrays (51)7 International Patent Classification(s) C12Q001/68 C07H 021/04 C07H 021/02 C12P 019/34 (21) Application No: 199875933 (22) Application Date: 1998.05.21 (87) WIPO No: WO98/53103 (30) Priority Data (31) Number (32) Date (33) Country 08/859998 1997.05.21 US 09/053375 1998.03.31 US (43) Publication Date : 1998.12.11 (43) Publication Journal Date : 1999.02.04 (44) Accepted Journal Date : 2001.12.20 (71) Applicant(s) Clontech Laboratories, Inc. (72) Inventor(s) Alex Chenchik; George Jokhadze; Robert Bibilashvilli (74) Agent/Attorney F.B. RICE and CO.,139 Rathdowne Street,CARLTON VIC 3053 (56) Related Art PROC NATL ACAD SCI USA 93,10614-9 ANCEL BIOCHEM 216,299-304 CRENE 156,207-13 OPI DAtE 11/12/98 APPLN. ID 75933/98 AOJP DATE 04/02/99 PCT NUMBER PCT/US98/10561 IIIIIIIUIIIIIIIIIIIIIIIIIIIII AU9875933 .PCT) (51) International Patent Classification 6 ; (11) International Publication Number: WO 98/53103 C12Q 1/68, C12P 19/34, C07H 2UO2, Al 21/04 (43) International Publication Date: 26 November 1998 (26.11.98) (21) International Application Number: PCT/US98/10561 (81) Designated States: AL, AM, AT, AU, AZ, BA, BB, BG, BR, BY, CA, CH, CN, CU, CZ, DE, DK, EE, ES, FI, GB, GE, (22) International Filing Date: 21 May 1998 (21.05.98) GH, GM, GW, HU, ID, IL, IS, JP, KE, KG, KP, KR, KZ, LC, LK, LR, LS, LT, LU, LV, MD, MG, MK, MN, MW, MX, NO, NZ, PL, PT, RO, RU, SD, SE, SG, SI, SK, SL, (30) Priority Data: TJ, TM, TR, TT, UA, UG, US, UZ, VN, YU, ZW, ARIPO 08/859,998 21 May 1997 (21.05.97) US patent (GH, GM, KE, LS, MW, SD, SZ, UG, ZW), Eurasian 09/053,375 31 March 1998 (31.03.98) US patent (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), European patent (AT, BE, CH, CY, DE, DK, ES, Fl, FR, GB, GR, IE, IT, LU, MC, NL, PT, SE), OAPI patent (BF, BJ, CF, (71) Applicant (for all designated States except US): CLONTECH CG, CI, CM, GA, GN, ML, MR, NE, SN, TD, TG). -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Electronic Supplementary Material (ESI) for Green Chemistry. This Journal Is © the Royal Society of Chemistry 2016
Electronic Supplementary Material (ESI) for Green Chemistry. This journal is © The Royal Society of Chemistry 2016 Electronic Supplementary Information for: Lignin depolymerization by fungal secretomes and a microbial sink† Davinia Salvachúaa,‡, Rui Katahiraa,‡, Nicholas S. Clevelanda, Payal Khannaa, Michael G. Rescha, Brenna A. Blacka, Samuel O. Purvineb, Erika M. Zinkb, Alicia Prietoc, María J. Martínezc, Angel T. Martínezc, Blake A. Simmonsd,e, John M. Gladdend,f, Gregg T. Beckhama,* a. National Bioenergy Center, National Renewable Energy Laboratory (NREL), Golden CO 80401, USA b. Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory (PNNL), Richland, WA 99352, USA c. Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas (CSIC), E-28040 Madrid, Spain d. Joint BioEnergy Institute (JBEI), Emeryville, CA 94608 e. Biological Systems and Engineering, Lawrence Berkeley National Laboratory, Berkeley CA 94720 USA f. Sandia National Laboratory, Livermore CA 94550 ‡ Equal contribution * Corresponding author: [email protected] Extension of materials and methods section Analysis of aromatics by LC-MS/MS Mass spectrometry was used in the last experiment of the current study to analyze aromatics from the soluble fraction. For this purpose, 14.5 mg of freeze-dried supernatant from 8 different treatments was reconstituted in 1 mL methanol. Analysis of samples was performed on an Agilent 1100 LC system equipped with a diode array detector (DAD) and an Ion Trap SL MS (Agilent Technologies, Palo Alto, CA) with in-line electrospray ionization (ESI). Each sample was injected at a volume of 25 μL into the LC-MS system. Primary degradation compounds were separated using a YMC C30 Carotenoid 0.3 μm, 4.6 x 150 mm column (YMC America, Allentown, PA) at an oven temperature of 30°C. -

Isolation and Genome Characterization of the Virulent Staphylococcus Aureus Bacteriophage SA97
Article Isolation and Genome Characterization of the Virulent Staphylococcus aureus Bacteriophage SA97 Yoonjee Chang 1, Hakdong Shin 1, Ju-Hoon Lee 2, Chul Jong Park 3, Soon-Young Paik 4 and Sangryeol Ryu 1,* Received: 21 July 2015 ; Accepted: 22 September 2015 ; Published: 1 October 2015 Academic Editor: Rob Lavigne 1 Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Sciences, and Center for Food and Bioconvergence, Seoul National University, Seoul 151-921, Korea; [email protected] (Y.C.); [email protected] (H.S.) 2 Department of Food Science and Biotechnology, Kyung Hee University, Yongin 446-701, Korea; [email protected] 3 Department of Dermatology, College of Medicine, the Catholic University of Korea, Seoul 137-701, Korea; [email protected] 4 Department of Microbiology, College of Medicine, the Catholic University of Korea, Seoul 137-701, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-2-880-4863; Fax: +82-2-873-5095 Abstract: A novel bacteriophage that infects S. aureus, SA97, was isolated and characterized. The phage SA97 belongs to the Siphoviridae family, and the cell wall teichoic acid (WTA) was found to be a host receptor of the phage SA97. Genome analysis revealed that SA97 contains 40,592 bp of DNA encoding 54 predicted open reading frames (ORFs), and none of these genes were related to virulence or drug resistance. Although a few genes associated with lysogen formation were detected in the phage SA97 genome, the phage SA97 produced neither lysogen nor transductant in S. aureus. -

On the Neurobiology of Physical Activity in Mice and Human in Search of Eating Disorders Determinants
On the Neurobiology of Physical Activity in Mice and Human In Search of Eating Disorders Determinants Elżbieta Kostrzewa ISBN 9789-0393-6090-3 Printed by: CPI Koninklijke Wöhrmann Layout: Elżbieta Kostrzewa and Wouter Wiltenburg Cover design: Elżbieta Kostrzewa and Wouter Wiltenburg More artwork at surrella.wordpress.com . This cover was inspired by The Elephant in the Room , Banksy exhibition, 2006 Barely Legal show, Los Angeles. © Elżbieta Kostrzewa On the Neurobiology of Physical Activity in Mice and Human In Search of Eating Disorders Determinants Over de Neurobiologie van Lichaamsbeweging in Muizen en Mens Op Zoek naar Determinanten van Eetstoornissen (met een samenvatting in het Nederlands) Badania nad neurobiologicznymi podstawami aktywności fizycznej myszy i ludzi W poszukiwaniu determinantów zaburzeń odżywiania (ze streszczeniem w języku polskim) Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht op gezag van de rector magnificus, prof.dr. G.J. van der Zwaan, ingevolge het besluit van het college voor promoties in het openbaar te verdedigen op donderdag 6 februari 2014 des ochtends te 10.30 uur door Elżbieta Kostrzewa geboren op 10 maart 1983 te Krakau, Polen Promotor: Prof. dr. R.A.H. Adan Co-promotor: Dr. M.J.H. Kas Table of Contents CHAPTER 1 07 General Introduction Physical Activity 08 Eating Disorders 29 Translational Approach in Genetic Studies 43 Scope of the Thesis 49 CHAPTER 2 53 A Candidate Syntenic Genetic Locus is Associated with Physical Activity Levels in Mice and Humans CHAPTER 3 77