Genetic Expression Profile Analysis of the Temporal Inhibition of Quercetin and Naringenin on Lactobacillus Rhamnosus GG
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genome of Nanoarchaeum Equitans: Insights Into Early Archaeal Evolution and Derived Parasitism
The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism Elizabeth Waters†‡, Michael J. Hohn§, Ivan Ahel¶, David E. Graham††, Mark D. Adams‡‡, Mary Barnstead‡‡, Karen Y. Beeson‡‡, Lisa Bibbs†, Randall Bolanos‡‡, Martin Keller†, Keith Kretz†, Xiaoying Lin‡‡, Eric Mathur†, Jingwei Ni‡‡, Mircea Podar†, Toby Richardson†, Granger G. Sutton‡‡, Melvin Simon†, Dieter So¨ ll¶§§¶¶, Karl O. Stetter†§¶¶, Jay M. Short†, and Michiel Noordewier†¶¶ †Diversa Corporation, 4955 Directors Place, San Diego, CA 92121; ‡Department of Biology, San Diego State University, 5500 Campanile Drive, San Diego, CA 92182; §Lehrstuhl fu¨r Mikrobiologie und Archaeenzentrum, Universita¨t Regensburg, Universita¨tsstrasse 31, D-93053 Regensburg, Germany; ‡‡Celera Genomics Rockville, 45 West Gude Drive, Rockville, MD 20850; Departments of ¶Molecular Biophysics and Biochemistry and §§Chemistry, Yale University, New Haven, CT 06520-8114; and ʈDepartment of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061 Communicated by Carl R. Woese, University of Illinois at Urbana–Champaign, Urbana, IL, August 21, 2003 (received for review July 22, 2003) The hyperthermophile Nanoarchaeum equitans is an obligate sym- (6–8). Genomic DNA was either digested with restriction en- biont growing in coculture with the crenarchaeon Ignicoccus. zymes or sheared to provide clonable fragments. Two plasmid Ribosomal protein and rRNA-based phylogenies place its branching libraries were made by subcloning randomly sheared fragments point early in the archaeal lineage, representing the new archaeal of this DNA into a high-copy number vector (Ϸ2.8 kbp library) kingdom Nanoarchaeota. The N. equitans genome (490,885 base or low-copy number vector (Ϸ6.3 kbp library). DNA sequence pairs) encodes the machinery for information processing and was obtained from both ends of plasmid inserts to create repair, but lacks genes for lipid, cofactor, amino acid, or nucleotide ‘‘mate-pairs,’’ pairs of reads from single clones that should be biosyntheses. -

Mullergliaregnerationtranscriptome
gene.id fc1 fc2 fc3 fc4 p1 p2 p3 p4 FDR.pvalue-1 Gene Symbol Gene Title Pathway go biological process term go molecular function term go cellular component term Dr.10016.1.A1_at -2.33397 -3.86923 -4.38335 -2.39965 0.935201 0.320614 0.208 0.917227 0.208 zgc:77556 zgc:77556 --- proteolysis arylesterase activity /// metallopeptidase activity /// zinc ion --- binding Dr.10024.1.A1 at 1.483417 2.531269 2.089091 1.698761 1 0.613 0.998961 1 0.613 zgc:171808 zgc:171808--- cell-cell signaling --- --- Dr.10051.1.A1 at -1.78449 -2.22024 -1.70922 -1.99464 1 0.901663 1 0.999955 0.9017 ccng2 cyclin G2 --- --- --- --- Dr.10061.1.A1 at 2.065955 2.274632 2.248958 2.507754 0.992 0.718655 0.83 0.600609 0.6006 zgc:173506 zgc:173506--- --- --- --- Dr.10061.2.A1 at 2.131883 2.616483 2.49378 2.815337 0.983443 0.711513 0.805599 0.519115 0.5191 zgc:173506 Zgc:17350 --- --- --- --- Dr.10065.1.A1 at -1.02315 -2.01596 -2.29343 -1.88944 1 0.999955 0.957199 1 0.9572 zgc:114139 zgc:114139--- --- --- cytoplasm /// centrosome Dr.10070.1.A1_at -1.74365 -2.52206 -2.39093 -1.86817 1 0.741254 0.885401 1 0.7413 fbp1a fructose-1, --- carbohydrate metabolic process hydrolase activity /// phosphoric ester hydrolase activity --- Dr.10074.1.S1_at 6.035545 10.44051 7.880519 5.020371 0.1104 0.044 0.062491 0.144679 0.044 pdgfaa platelet-der--- multicellular organismal development /// cell proliferation growth factor activity membrane Dr.10095.1.A1 at -1.73408 -2.11615 -1.47234 -2.19919 1 0.997562 1 0.978177 0.9782 wu:fk95g04 wu:fk95g04--- --- --- --- Dr.10110.1.S1 a at 3.929761 5.798708 -

1/05661 1 Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date _ . ... - 12 May 2011 (12.05.2011) W 2 11/05661 1 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12Q 1/00 (2006.0 1) C12Q 1/48 (2006.0 1) kind of national protection available): AE, AG, AL, AM, C12Q 1/42 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) Number: International Application DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/US20 10/054171 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 26 October 2010 (26.10.2010) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, (25) Filing Language: English SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, (26) Publication Language: English TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/255,068 26 October 2009 (26.10.2009) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, (71) Applicant (for all designated States except US): ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, MYREXIS, INC. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

AUSTRALIAN PATENT OFFICE (11) Application No. AU 199875933 B2
(12) PATENT (11) Application No. AU 199875933 B2 (19) AUSTRALIAN PATENT OFFICE (10) Patent No. 742342 (54) Title Nucleic acid arrays (51)7 International Patent Classification(s) C12Q001/68 C07H 021/04 C07H 021/02 C12P 019/34 (21) Application No: 199875933 (22) Application Date: 1998.05.21 (87) WIPO No: WO98/53103 (30) Priority Data (31) Number (32) Date (33) Country 08/859998 1997.05.21 US 09/053375 1998.03.31 US (43) Publication Date : 1998.12.11 (43) Publication Journal Date : 1999.02.04 (44) Accepted Journal Date : 2001.12.20 (71) Applicant(s) Clontech Laboratories, Inc. (72) Inventor(s) Alex Chenchik; George Jokhadze; Robert Bibilashvilli (74) Agent/Attorney F.B. RICE and CO.,139 Rathdowne Street,CARLTON VIC 3053 (56) Related Art PROC NATL ACAD SCI USA 93,10614-9 ANCEL BIOCHEM 216,299-304 CRENE 156,207-13 OPI DAtE 11/12/98 APPLN. ID 75933/98 AOJP DATE 04/02/99 PCT NUMBER PCT/US98/10561 IIIIIIIUIIIIIIIIIIIIIIIIIIIII AU9875933 .PCT) (51) International Patent Classification 6 ; (11) International Publication Number: WO 98/53103 C12Q 1/68, C12P 19/34, C07H 2UO2, Al 21/04 (43) International Publication Date: 26 November 1998 (26.11.98) (21) International Application Number: PCT/US98/10561 (81) Designated States: AL, AM, AT, AU, AZ, BA, BB, BG, BR, BY, CA, CH, CN, CU, CZ, DE, DK, EE, ES, FI, GB, GE, (22) International Filing Date: 21 May 1998 (21.05.98) GH, GM, GW, HU, ID, IL, IS, JP, KE, KG, KP, KR, KZ, LC, LK, LR, LS, LT, LU, LV, MD, MG, MK, MN, MW, MX, NO, NZ, PL, PT, RO, RU, SD, SE, SG, SI, SK, SL, (30) Priority Data: TJ, TM, TR, TT, UA, UG, US, UZ, VN, YU, ZW, ARIPO 08/859,998 21 May 1997 (21.05.97) US patent (GH, GM, KE, LS, MW, SD, SZ, UG, ZW), Eurasian 09/053,375 31 March 1998 (31.03.98) US patent (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), European patent (AT, BE, CH, CY, DE, DK, ES, Fl, FR, GB, GR, IE, IT, LU, MC, NL, PT, SE), OAPI patent (BF, BJ, CF, (71) Applicant (for all designated States except US): CLONTECH CG, CI, CM, GA, GN, ML, MR, NE, SN, TD, TG). -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Electronic Supplementary Material (ESI) for Green Chemistry. This Journal Is © the Royal Society of Chemistry 2016
Electronic Supplementary Material (ESI) for Green Chemistry. This journal is © The Royal Society of Chemistry 2016 Electronic Supplementary Information for: Lignin depolymerization by fungal secretomes and a microbial sink† Davinia Salvachúaa,‡, Rui Katahiraa,‡, Nicholas S. Clevelanda, Payal Khannaa, Michael G. Rescha, Brenna A. Blacka, Samuel O. Purvineb, Erika M. Zinkb, Alicia Prietoc, María J. Martínezc, Angel T. Martínezc, Blake A. Simmonsd,e, John M. Gladdend,f, Gregg T. Beckhama,* a. National Bioenergy Center, National Renewable Energy Laboratory (NREL), Golden CO 80401, USA b. Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory (PNNL), Richland, WA 99352, USA c. Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas (CSIC), E-28040 Madrid, Spain d. Joint BioEnergy Institute (JBEI), Emeryville, CA 94608 e. Biological Systems and Engineering, Lawrence Berkeley National Laboratory, Berkeley CA 94720 USA f. Sandia National Laboratory, Livermore CA 94550 ‡ Equal contribution * Corresponding author: [email protected] Extension of materials and methods section Analysis of aromatics by LC-MS/MS Mass spectrometry was used in the last experiment of the current study to analyze aromatics from the soluble fraction. For this purpose, 14.5 mg of freeze-dried supernatant from 8 different treatments was reconstituted in 1 mL methanol. Analysis of samples was performed on an Agilent 1100 LC system equipped with a diode array detector (DAD) and an Ion Trap SL MS (Agilent Technologies, Palo Alto, CA) with in-line electrospray ionization (ESI). Each sample was injected at a volume of 25 μL into the LC-MS system. Primary degradation compounds were separated using a YMC C30 Carotenoid 0.3 μm, 4.6 x 150 mm column (YMC America, Allentown, PA) at an oven temperature of 30°C. -

Isolation and Genome Characterization of the Virulent Staphylococcus Aureus Bacteriophage SA97
Article Isolation and Genome Characterization of the Virulent Staphylococcus aureus Bacteriophage SA97 Yoonjee Chang 1, Hakdong Shin 1, Ju-Hoon Lee 2, Chul Jong Park 3, Soon-Young Paik 4 and Sangryeol Ryu 1,* Received: 21 July 2015 ; Accepted: 22 September 2015 ; Published: 1 October 2015 Academic Editor: Rob Lavigne 1 Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Sciences, and Center for Food and Bioconvergence, Seoul National University, Seoul 151-921, Korea; [email protected] (Y.C.); [email protected] (H.S.) 2 Department of Food Science and Biotechnology, Kyung Hee University, Yongin 446-701, Korea; [email protected] 3 Department of Dermatology, College of Medicine, the Catholic University of Korea, Seoul 137-701, Korea; [email protected] 4 Department of Microbiology, College of Medicine, the Catholic University of Korea, Seoul 137-701, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-2-880-4863; Fax: +82-2-873-5095 Abstract: A novel bacteriophage that infects S. aureus, SA97, was isolated and characterized. The phage SA97 belongs to the Siphoviridae family, and the cell wall teichoic acid (WTA) was found to be a host receptor of the phage SA97. Genome analysis revealed that SA97 contains 40,592 bp of DNA encoding 54 predicted open reading frames (ORFs), and none of these genes were related to virulence or drug resistance. Although a few genes associated with lysogen formation were detected in the phage SA97 genome, the phage SA97 produced neither lysogen nor transductant in S. aureus. -

Molecular Landscapes of Deoxyuridine 5'-Triphosphatase (Dutpase) As a Drug Target Against Camel Trypanosoma Evansi
Pakistan Veterinary Journal ISSN: 0253-8318 (PRINT), 2074-7764 (ONLINE) DOI: 10.29261/pakvetj/2021.015 RESEARCH ARTICLE Molecular Landscapes of Deoxyuridine 5'-Triphosphatase (dUTPase) as a Drug Target against Camel Trypanosoma evansi Mahmoud Kandeel1,2* and Abdulla Al-Taher1 1Department of Biomedical Sciences, College of Veterinary Medicine, King Faisal University, Al-Ahsa, 31982, Saudi Arabia; 2Department of Pharmacology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafrelsheikh, 35255, Egypt *Corresponding author: [email protected] ARTICLE HISTORY (20-562) ABSTRACT Received: October 29, 2020 The decoding of genome sequences of camel and its pathogens will help in speeding Revised: December 18, 2020 up the discovery of new drug targets of pathogens. In this context, Trypanosoma Accepted: December 19, 2020 Published online: January 24, 2021 evansi (T. evansi) constitutes the major health hazard with confirmed broad host Key words: range and zoonotic infections of humans. Discovery of a drug target in T. evansi Camel deoxyuridine 5'- triphosphate (dUTP) pathways by comparing dUTP metabolizing drug discovery enzymes in dromedary camels and the parasite. The pyrimidine pathways were dUTP investigated and the enzymes involved in the metabolism of deoxyuridine 5'- dUTPase triphosphate (dUTP) in camel and T. evansi were investigated by bioinformatics Pyrimidine tools. T. evansi was devoid of deoxycytidine triphosphate deaminase rendering the Trypanosoma evansi source of dUTP under metabolic stress. There were interesting differences in predicted structure and function of dUTPase between the camel and T. evansi. The camel dUTPase is a trimeric enzyme of 165 amino acids highly similar to other vertebrates’ enzyme, mw of 18 kDa and form trimers without multiple domain constituents. -

Title Proteomic Approach to Study the Cytotoxicity of Dioscin
CORE Metadata, citation and similar papers at core.ac.uk Provided by HKU Scholars Hub Proteomic approach to study the cytotoxicity of dioscin Title (saponin) Author(s) Wang, Y; Yim, HC; Yang, Z; Chiu, JF; Che, CM; He, QY Citation Proteomics, 2006, v. 6 n. 8, p. 2422-2432 Issued Date 2006 URL http://hdl.handle.net/10722/48497 Rights Creative Commons: Attribution 3.0 Hong Kong License This is a pre-published version Proteomic approach to study the cytotoxicity of dioscin (saponin) Ying Wang1, Yim Hing Cheung2, Zhiqi Yang1,3, Jen-Fu Chiu2, Chi-Ming Che1*, and Qing-Yu He1* 1Department of Chemistry and Open Laboratory of Chemical Biology, 2Department of Anatomy, University of Hong Kong, Pokfulam, Hong Kong, China 3Shanghai Institute of Organic Chemistry, Shanghai, China Running title: Proteomics of Dioscin Cytotoxicity Correspondence to: Dr. Qing-Yu He, Department of Chemistry, University of Hong Kong, Pokfulam, Hong Kong, China. Tel: (852)2299-0787, Fax: (852)2817-1006, E-mail: [email protected] Or: Prof. Chi-Ming Che, Department of Chemistry, University of Hong Kong, Pokfulam, Hong Kong, China. Fax: (852)2857-1586, E-mail: [email protected] List of Abbreviations: DAPI, 4,6-diamidino-2-phenylindole; IC50, half-maximal inhibitory concentration; MALDI- TOF MS, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; ∆Ψm, mitochondrial transmembrane potential; PMF, peptide mass fingerprinting; PI, propidium idodide; Hsp, Heat shock protein ; Rho GDI, Rho GDP-dissociation inhibitor; PTPase, protein- tyrosine phosphatase; dUTPase, dUTP diphosphatase; PPase, inorganic pyrophosphatase; hnRNP, heterogeneous nuclear ribonucleoprotein; HEI-C, human enhancer of invasion-cluster. 1 Abstract Dioscin, extracted from the root of Polygonatum Zanlanscianense pamp, exhibits cytotoxicity towards human myeloblast leukemia HL-60 cells. -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine -
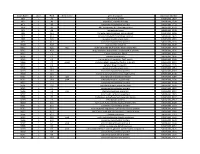
NTML 384-Well Array Well Identification
Strain Name plate Well Gene name gene discription Accession Number NE1 1 A1 cell surface protein SAUSA300_1327 NE2 1 A3 peptidase, rhomboid family SAUSA300_1509 NE3 1 A5 conserved hypothetical protein SAUSA300_1733 NE4 1 A7 ABC transporter ATP-binding protein SAUSA300_0309 NE5 1 A9 aminotransferase SAUSA300_2539 NE6 1 A11 formate dehydrogenase, alpha subunit SAUSA300_2258 NE7 1 A13 conserved hypothetical phage protein SAUSA300_1425 NE8 1 A15 putative membrane protein SAUSA300_1481 NE9 1 A17 conserved hypothetical protein SAUSA300_1294 NE10 1 A19 putative hemolysin III SAUSA300_2129 NE11 1 A21 recJ single-stranded-DNA-specific exonuclease RecJ SAUSA300_1592 NE12 1 A23 drug resistance transporter, EmrB/QacA subfamily SAUSA300_2126 NE13 1 C1 ribose transporter RbsU SAUSA300_0264 NE14 1 C3 putative transporter SAUSA300_2406 NE15 1 C5 transcriptional regulator, TetR family SAUSA300_2509 NE16 1 C7 moaD molybdopterin converting factor, subunit 1 SAUSA300_2221 NE17 1 C9 putative drug transporter SAUSA300_1705 NE18 1 C11 putative membrane protein SAUSA300_1851 NE19 1 C13 conserved hypothetical protein SAUSA300_0369 NE20 1 C15 transcriptional antiterminator, BglG family SAUSA300_0238 NE21 1 C17 upp uracil phosphoribosyltransferase SAUSA300_2066 NE22 1 C19 polA DNA polymerase I superfamily SAUSA300_1636 NE23 1 C21 conserved hypothetical protein SAUSA300_0740 NE24 1 C23 secretory antigen precursor SsaA SAUSA300_2503 NE25 1 E1 conserved hypothetical protein SAUSA300_1905 NE26 1 E3 coa staphylocoagulase precursor SAUSA300_0224 NE27 1 E5 nitric oxide