Bacterial Communities of Aphis Gossypii and Myzus Persicae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Check List Notes on Geographic Distribution Check List 13(2): 2091, 12 April 2017 Doi: ISSN 1809-127X © 2017 Check List and Authors
13 2 2091 the journal of biodiversity data 12 April 2017 Check List NOTES ON GEOGRAPHIC DISTRIBUTION Check List 13(2): 2091, 12 April 2017 doi: https://doi.org/10.15560/13.2.2091 ISSN 1809-127X © 2017 Check List and Authors First records of Sturnira bakeri Velazco & Patterson, 2014 (Chiroptera: Phyllostomidae) from Colombia Sebastián Montoya-Bustamante1, 3, Baltazar González-Chávez2, Natalya Zapata-Mesa1 & Laura Obando-Cabrera1 1 Universidad del Valle, Grupo de Investigación en Ecología Animal, Departamento de Biología, Apartado Aéreo 25360, Cali, Colombia 2 Universidad del Valle, Apartado Aéreo 25360, Cali, Colombia 3 Corresponding author. E-mail: [email protected] Abstract: We evaluate the occurrence of S. bakeri in m). Therefore, there was a high probability that this species Colombia, a recently described species. We report seven inhabits northwestern Peru rather than elsewhere (Velaz- new records and include data on skull measurements of co & Patterson 2014). Recently, Sánchez & Pacheco these individuals and information on the new localities. A (2016) tested this hypothesis and reported the presence of discriminant analysis suggests that condyloincisive length S. bakeri from six localities in northwestern Peru. No fur- and dentary length are the most important measurements ther information has been published on S. bakeri and its to separate S. bakeri and S. luisi from S. lilium. However, geographic distribution remains obscure. Our aim was to to distinguish S. bakeri from S. luisi, we used discrete evaluate the occurrence of S. bakeri in Colombia, as well as characters proposed in the original descriptions of these to increase our knowledge of the morphological variation two taxa. -

New Insights Into the Microbiota of Moth Pests
International Journal of Molecular Sciences Review New Insights into the Microbiota of Moth Pests Valeria Mereghetti, Bessem Chouaia and Matteo Montagna * ID Dipartimento di Scienze Agrarie e Ambientali, Università degli Studi di Milano, 20122 Milan, Italy; [email protected] (V.M.); [email protected] (B.C.) * Correspondence: [email protected]; Tel.: +39-02-5031-6782 Received: 31 August 2017; Accepted: 14 November 2017; Published: 18 November 2017 Abstract: In recent years, next generation sequencing (NGS) technologies have helped to improve our understanding of the bacterial communities associated with insects, shedding light on their wide taxonomic and functional diversity. To date, little is known about the microbiota of lepidopterans, which includes some of the most damaging agricultural and forest pests worldwide. Studying their microbiota could help us better understand their ecology and offer insights into developing new pest control strategies. In this paper, we review the literature pertaining to the microbiota of lepidopterans with a focus on pests, and highlight potential recurrent patterns regarding microbiota structure and composition. Keywords: symbiosis; bacterial communities; crop pests; forest pests; Lepidoptera; next generation sequencing (NGS) technologies; diet; developmental stages 1. Introduction Insects represent the most successful taxa of eukaryotic life, being able to colonize almost all environments, including Antarctica, which is populated by some species of chironomids (e.g., Belgica antarctica, Eretmoptera murphyi, and Parochlus steinenii)[1,2]. Many insects are beneficial to plants, playing important roles in seed dispersal, pollination, and plant defense (by feeding upon herbivores, for example) [3]. On the other hand, there are also damaging insects that feed on crops, forest and ornamental plants, or stored products, and, for these reasons, are they considered pests. -

Evaluation of Nama Opportunities in Colombia's
EVALUATION OF NAMA OPPORTUNITIES IN COLOMBIA’S SOLID WASTE SECTOR WRITTEN BY: Leo Larochelle Michael Turner Michael LaGiglia CCAP RESEARCH SUPPORT: CENTER FOR CLEAN AIR POLICY Hill Consulting (Bogotá) OCTOBER 2012 Dialogue. Insight. Solutions. Acknowledgements This paper is a product of CCAP’s Mitigation Action Implementation Network (MAIN) and was written by Leo Larochelle, Michael Turner, and Michael LaGiglia of CCAP. This project was undertaken with the financial support of the Government of Canada through the Federal Department of the Environment. Special thanks are due to the individuals and organizations in Colombia who offered their time and assistance, through phone interviews or in-person discussions to help inform this work. The support of the Ministerio de Ambiente y Desarrollo Sostenible was essential to the success of this report as well as help from the Steering Committee (made up of the Ministerio de Ambiente Vivienda Y Desarrollo Territorial, the Departamento Nacional de Planeación, the Ministerio de Ambiente y Desarrollo Sostenible, and the Superintendencia de Servicios Públicos Domiciliarios), representatives from Santiago de Cali (Empresa Pública de Gestión Integral de Residuos Sólidos de Cali, Departamento Administrativo para la Gestión del Medio Ambiente), Medellín (Area Metropolitana del Valle de Aburra Unidad Ambiental), Ibagué (Corporación Autónoma Regional del Tolima-Cortolima and Interaseo) and Sogamoso (Secretario de Desarrollo y Medio Ambiente and Coservicios). The views expressed in this paper represent those -

Biodiversity Climate Change Impacts Report Card Technical Paper 12. the Impact of Climate Change on Biological Phenology In
Sparks Pheno logy Biodiversity Report Card paper 12 2015 Biodiversity Climate Change impacts report card technical paper 12. The impact of climate change on biological phenology in the UK Tim Sparks1 & Humphrey Crick2 1 Faculty of Engineering and Computing, Coventry University, Priory Street, Coventry, CV1 5FB 2 Natural England, Eastbrook, Shaftesbury Road, Cambridge, CB2 8DR Email: [email protected]; [email protected] 1 Sparks Pheno logy Biodiversity Report Card paper 12 2015 Executive summary Phenology can be described as the study of the timing of recurring natural events. The UK has a long history of phenological recording, particularly of first and last dates, but systematic national recording schemes are able to provide information on the distributions of events. The majority of data concern spring phenology, autumn phenology is relatively under-recorded. The UK is not usually water-limited in spring and therefore the major driver of the timing of life cycles (phenology) in the UK is temperature [H]. Phenological responses to temperature vary between species [H] but climate change remains the major driver of changed phenology [M]. For some species, other factors may also be important, such as soil biota, nutrients and daylength [M]. Wherever data is collected the majority of evidence suggests that spring events have advanced [H]. Thus, data show advances in the timing of bird spring migration [H], short distance migrants responding more than long-distance migrants [H], of egg laying in birds [H], in the flowering and leafing of plants[H] (although annual species may be more responsive than perennial species [L]), in the emergence dates of various invertebrates (butterflies [H], moths [M], aphids [H], dragonflies [M], hoverflies [L], carabid beetles [M]), in the migration [M] and breeding [M] of amphibians, in the fruiting of spring fungi [M], in freshwater fish migration [L] and spawning [L], in freshwater plankton [M], in the breeding activity among ruminant mammals [L] and the questing behaviour of ticks [L]. -

Anatomical Investigations of the Male Reproductive System of Selected Species of Macrosiphini
Bulletin of Insectology 61 (1): 179, 2008 ISSN 1721-8861 Anatomical investigations of the male reproductive system of selected species of Macrosiphini Karina WIECZOREK Department of Zoology, Faculty of Biology and Environmental Protection, University of Silesia, Katowice, Poland Abstract Histological sections and whole mount preparations of five species of Macrosiphini [Impatientinum asiaticum Nevsky, Hypero- myzus (Hyperomyzus) pallidus Hille Ris Lambers, Myzus (Myzus) cerasi (F.), Rhopalomyzus (Judenkoa) loniceare (Siebold) and Uroleucon obscurum (Koch)] were examined. Key words: Hemiptera, Aphidoidea, Aphididae, Macrosiphini, male reproductive system. In previous research on the structure of the male repro- References ductive system of aphids, about 70 species from various subfamilies have been described, mainly Lachninae BLACKMAN R. L., 1987.- Reproduction cytogenetics and de- (Wojciechowski, 1977), Chaitophorinae (Wieczorek and velopment, pp 163-191. In: Aphids, their biology, natural Wojciechowski, 2004), and Calaphidinae (Głowacka et. enemies and control (MINKS A. K., HARREWIJN P., Ed).- El- sevier, Amsterdam, The Netherland. al., 1974; Wieczorek and Wojciechowski, 2001; Wiec- BOCHEN K., KLIMASZEWSKI S. M., WOJCIECHOWSKI W., zorek, 2006). 1975.- Budowa męskiego układu rozrodczego Macrosipho- In contrast, Aphidinae are the largest and most diverse niella artemisiae (B.De Fonsc.) i M. millefolli (De Geer) group of aphids whose male reproductive system is least (Homoptera, Aphididae).- Acta Biologica Uniwersytet Slaski studied. In Pterocommatini the structure of the male re- w Katowicach, 90: 73-81. productive system has been analysed in Pterocomma GŁOWACKA E., KLIMASZEWSKI S. M., SZELEGIEWICZ H., WOJ- populeum (Kaltenbach) (Wieczorek and Wo- CIECHOWSKI W., 1974.- Uber den Bau des mannlichen Fort- jciechowski, 2005) and Pterocomma salicis (L.) (Wiec- pflanzungssystems der Aphiden (Homoptera, Aphidoidea).- zorek and Mróz, 2006), in Aphidini in Rhopalosiphum Annales Universitas Mariae Curie-Skłodowska, 29C: 133-138. -

Effects of Habitat Fragmentation and Disturbance on Biodiversity and Ecosystem Functions
Effects of habitat fragmentation and disturbance on biodiversity and ecosystem functions Inauguraldissertation der Philosophisch-naturwissenschaftlichen Fakultät der Universität Bern vorgelegt von Christof Schüepp von Eschlikon TG Leiter der Arbeit: Prof. Dr. M. H. Entling Institut für Ökologie und Evolution | downloaded: 13.3.2017 Von der Philosophisch-naturwissenschaftlichen Fakultät angenommen. Bern, 14. Mai 2013 Der Dekan: Prof. Dr. S. Decurtins Originaldokument gespeichert auf dem Webserver der Universitätsbibliothek Bern Dieses Werk ist unter einem https://doi.org/10.7892/boris.54819 Creative Commons Namensnennung-Keine kommerzielle Nutzung-Keine Bearbeitung 2.5 Schweiz Lizenzvertrag lizenziert. Um die Lizenz anzusehen, gehen Sie bitte zu http://creativecommons.org/licenses/by-nc-nd/2.5/ch/ oder schicken Sie einen Brief an Creative Commons, 171 Second Street, Suite 300, San Francisco, California 94105, USA. source: Effects of habitat fragmentation and disturbance on biodiversity and ecosystem functions Creative Commons Licence Urheberrechtlicher Hinweis Dieses Dokument steht unter einer Lizenz der Creative Commons Namensnennung- Keine kommerzielle Nutzung-Keine Bearbeitung 2.5 Schweiz. http://creativecommons.org/licenses/by-nc-nd/2.5/ch/ Sie dürfen: dieses Werk vervielfältigen, verbreiten und öffentlich zugänglich machen Zu den folgenden Bedingungen: Namensnennung. Sie müssen den Namen des Autors/Rechteinhabers in der von ihm festgelegten Weise nennen (wodurch aber nicht der Eindruck entstehen darf, Sie oder die Nutzung des Werkes durch Sie würden entlohnt). Keine kommerzielle Nutzung. Dieses Werk darf nicht für kommerzielle Zwecke verwendet werden. Keine Bearbeitung. Dieses Werk darf nicht bearbeitet oder in anderer Weise verändert werden. Im Falle einer Verbreitung müssen Sie anderen die Lizenzbedingungen, unter welche dieses Werk fällt, mitteilen. Jede der vorgenannten Bedingungen kann aufgehoben werden, sofern Sie die Einwilligung des Rechteinhabers dazu erhalten. -
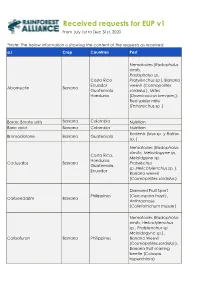
IIS 2020 Requests Overview
Received requests for EUP v1 From July 1st to Dec 31st, 2020 *Note: The below information is showing the content of the requests as received. a.i Crop Countries Pest Nematodes ( Radophulus similis, Radopholus sp, Costa Rica Pratylenchus sp ), Banana Ecuador weevil ( Cosmopolites Abamectin Banana Guatemala sordidus ), Mites Honduras (Dysmicoccus brevipes ), Red spider mite (Tetranychus sp .) Borax; Borate salts Banana Colombia Nutrition Boric acid Banana Colombia Nutrition Rodents ( Mus sp. y Rattus Bromadiolone Banana Guatemala sp. ) Nematodes ( Radopholus silmillis, Meloidogyne sp, Costa Rica, Meloidgune sp, Honduras, Cadusafos Banana Pratylechus Guatemala, sp.,Helicotylenchus sp. ), Ecuador Banana weevil (Cosmopolites sordidus ) Diamond Fruit Sport Philippines (Cercospora hayii ), Carbendazim Banana Anthracnose (Colletotrichum musae ) Nematod es (Radopholus similis, Helicotylenchus sp., Pratylenchus sp. Meloidogyne sp.) , Carbofuran Banana Philippines Banana Weevil (Cosmopolites sordidus ), Banana fruit scarring beetle (Colaspis hyperchlora) a.i Crop Countries Pest Black Sigatoka Ecuador, Costa (Mycosphaerella fijiensis ), Rica, Philippines, Yellow Sigatoka Chlorothalonil Banana Honduras, (Mycosphaerella Guatemala, musicola ), Banana Colombia Freckle ( Phyllosticta musarum ) Mealybug ( Dysmicoccus brevipes, Pseudococcus elisae, Pseudococcus sp, Ferrista sp ), Dysmicoccus sp , Scale insects (Aspidiotus destructor, Diaspis boisduvalii ), Thrips (Thrips florum, Franckliniella spp, Chaetanaphothrips signipennis ), Banana fruit Philippines, -

A Contribution to the Aphid Fauna of Greece
Bulletin of Insectology 60 (1): 31-38, 2007 ISSN 1721-8861 A contribution to the aphid fauna of Greece 1,5 2 1,6 3 John A. TSITSIPIS , Nikos I. KATIS , John T. MARGARITOPOULOS , Dionyssios P. LYKOURESSIS , 4 1,7 1 3 Apostolos D. AVGELIS , Ioanna GARGALIANOU , Kostas D. ZARPAS , Dionyssios Ch. PERDIKIS , 2 Aristides PAPAPANAYOTOU 1Laboratory of Entomology and Agricultural Zoology, Department of Agriculture Crop Production and Rural Environment, University of Thessaly, Nea Ionia, Magnesia, Greece 2Laboratory of Plant Pathology, Department of Agriculture, Aristotle University of Thessaloniki, Greece 3Laboratory of Agricultural Zoology and Entomology, Agricultural University of Athens, Greece 4Plant Virology Laboratory, Plant Protection Institute of Heraklion, National Agricultural Research Foundation (N.AG.RE.F.), Heraklion, Crete, Greece 5Present address: Amfikleia, Fthiotida, Greece 6Present address: Institute of Technology and Management of Agricultural Ecosystems, Center for Research and Technology, Technology Park of Thessaly, Volos, Magnesia, Greece 7Present address: Department of Biology-Biotechnology, University of Thessaly, Larissa, Greece Abstract In the present study a list of the aphid species recorded in Greece is provided. The list includes records before 1992, which have been published in previous papers, as well as data from an almost ten-year survey using Rothamsted suction traps and Moericke traps. The recorded aphidofauna consisted of 301 species. The family Aphididae is represented by 13 subfamilies and 120 genera (300 species), while only one genus (1 species) belongs to Phylloxeridae. The aphid fauna is dominated by the subfamily Aphidi- nae (57.1 and 68.4 % of the total number of genera and species, respectively), especially the tribe Macrosiphini, and to a lesser extent the subfamily Eriosomatinae (12.6 and 8.3 % of the total number of genera and species, respectively). -

Fase De Aprestamiento – Informe Ejecutivo
FORMULACIÓN DEL PLAN DE ORDENACIÓN Y MANEJO DE LA SUBZONA HIDROGRÁFICA 2631 CONTRATO CVC 650 DE 2017 SUPERVISOR CVC: Carolina Zúñiga S. - Flor Inés Marín Dirección de Planeación REPRESENTANTE LEGAL FUNDACIÓN PROAGUA: John Jairo Daza Basto Santiago de Cali Diciembre 20 de 2018 Plan de Ordenación y Manejo de la Subzona Hidrográfica 2631: Arroyohondo, Yumbo, Mulaló, Vijes, Yotoco, Mediacanoa y Piedras Fase de Aprestamiento – Informe Ejecutivo TABLA DE CONTENIDO INTRODUCCIÓN ................................................................................................................................ 9 LOCALIZACIÓN DE LA SUBZONA HIDROGRÁFICA ..................................................... 11 IDENTIFICACIÓN, CARACTERIZACIÓN Y PRIORIZACIÓN DE ACTORES ............ 13 ESTRATEGIA DE PARTICIPACIÓN ....................................................................................... 22 3.1. ESTRATEGIA PARA LA DIFUSIÓN Y COMUNICACIÓN ............................................................... 29 3.2. CONFORMACIÓN Y APOYO AL FUNCIONAMIENTO DEL CONSEJO DE CUENCA ............. 29 3.3. PARTICIPACIÓN EN EL PROCESO DE CONSULTA PREVIA ........................................................ 30 RECOPILACIÓN Y ANÁLISIS DE INFORMACIÓN EXISTENTE ................................... 32 ANÁLISIS SITUACIONAL INICIAL ....................................................................................... 36 5.1. COMPONENTE ABIÓTICO ................................................................................................................... 36 5.2. -

Reporte No. 058 | 16 - 31 Octubre De 2019
6/11/2019 Correo de PROPACIFICO - Contexto Pacífico No. 058 | 16 - 31 octubre 2019 Reporte No. 058 | 16 - 31 octubre de 2019 - Gremios y Gobernación del Valle continúan exigiendo al Invías el mantenimiento vial del departamento. La directora de la Cámara Colombiana de la Infraestructura (CCI) expresó al Ministerio de Transporte y al Invías la necesidad de intervenir la malla vial e incluir el presupuesto para las reparaciones pendientes de hace más de dos años. Ante esto, el director del Invías manifestó que recientemente se adjudicó un contrato de mantenimiento y el próximo año se invertirán cerca de $200 mil millones de pesos en la construcción de la vía Buga - Buenaventura (Ver más). Por su parte la gobernadora del Valle envió una carta al presidente Duque detallando el estado de las vías y su preocupación frente al tema. Ver más - Nueva aerolínea de bajo costo llega al aeropuerto de Cali. Se trata de JetSmart, una aerolínea de bajo costo que anunció el pasado 16 de octubre su llegada a Cali con vuelos directos hacia Santiago de Chile. La Aeronáutica Civil aprobó la operación de esta ruta, con lo cual Colombia tendría dos rutas de vuelos directos a Chile desde Bogotá y Cali con frecuencias semanales desde los $500.000 pesos que empezarán a partir del 20 de diciembre de este año. Se espera un demanda de cerca de 30.000 pasajeros al año en esta nueva ruta y, de alcanzar esta demanda, implicaría un fortalecimiento de las relaciones entre Cali y la Región Pacífico con Chile como parte de la Alianza del Pacífico. -

The Effects of Climate Change on the Pests and Diseases of Coffee
y & W log ea to th a e im r l F o Journal of Climatology & Weather C f r e o c l a a s n Groenen, Climatol Weather Forecasting 2018, 6:3 t r i n u g o J Forecasting DOI: 10.4172/2332-2594.1000239 ISSN: 2332-2594 Review Article Open Access The Effects of Climate Change on the Pests and Diseases of Coffee Crops in Mesoamerica Danielle Groenen* Department of Earth, Ocean and Atmospheric Science and Center for Ocean–Atmospheric Prediction Studies, Florida State University, Florida, United States *Corresponding author: Danielle Groenen, Department of Earth, Ocean and Atmospheric Science and Center for Ocean–Atmospheric Prediction Studies, Florida State University, Florida, United States, Tel: 18506442525; E-mail: [email protected] Received Date: August 30, 2018; Accepted Date: September 21, 2018; Published Date: September 26, 2018 Copyright: © 2018 Groenen D. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Coffee is an in-demand commodity that is being threatened by climate change. Increasing temperatures and rainfall variability are predicted in the region of Mexico and Central America (Mesoamerica). This region is plagued with pests and diseases that have already caused millions of dollars in damages and losses to the coffee industry. This paper examines three pests that negatively affect coffee plants: the coffee borer beetle, the black twig borer, and nematodes. In addition, this paper examines three diseases that can destroy coffee crops: bacterial blight, coffee berry disease, and coffee leaf rust. -

Coffee Plant the Coffee Plant Makes a Great Indoor, Outdoor Shade, Or Office Plant
Coffee Plant The coffee plant makes a great indoor, outdoor shade, or office plant. Water when dry or the plant will let you know when it droops. Do not let it sit in water so tip over the pot if you over water the plant. Preform the finger test to check for dryness. When the plant is dry about an inch down, water thoroughly. The plant will stay pot bound about two years at which time you will transplant and enjoy a beautiful ornamental plant. See below. Coffea From Wikipedia, the free encyclopedia This article is about the biology of coffee. For the beverage, see Coffee. Coffea Coffea arabica trees in Brazil Scientific classification Kingdom: Plantae (unranked): Angiosperms (unranked): Eudicots (unranked): Asterids Order: Gentianales Family: Rubiaceae Subfamily: Ixoroideae Tribe: Coffeeae[1] Genus: Coffea L. Type species Coffea arabica L.[2] Species Coffea ambongensis Coffea anthonyi Coffea arabica - Arabica Coffee Coffea benghalensis - Bengal coffee Coffea boinensis Coffea bonnieri Coffea canephora - Robusta coffee Coffea charrieriana - Cameroonian coffee - caffeine free Coffea congensis - Congo coffee Coffea dewevrei - Excelsa coffee Coffea excelsa - Liberian coffee Coffea gallienii Coffea liberica - Liberian coffee Coffea magnistipula Coffea mogeneti Coffea stenophylla - Sierra Leonian coffee Coffea canephora green beans on a tree in Goa, India. Coffea is a large genus (containing more than 90 species)[3] of flowering plants in the madder family, Rubiaceae. They are shrubs or small trees, native to subtropical Africa and southern Asia. Seeds of several species are the source of the popular beverage coffee. After their outer hull is removed, the seeds are commonly called "beans".