Wo 2011/022435 A2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Insights Into the Microbiota of Moth Pests
International Journal of Molecular Sciences Review New Insights into the Microbiota of Moth Pests Valeria Mereghetti, Bessem Chouaia and Matteo Montagna * ID Dipartimento di Scienze Agrarie e Ambientali, Università degli Studi di Milano, 20122 Milan, Italy; [email protected] (V.M.); [email protected] (B.C.) * Correspondence: [email protected]; Tel.: +39-02-5031-6782 Received: 31 August 2017; Accepted: 14 November 2017; Published: 18 November 2017 Abstract: In recent years, next generation sequencing (NGS) technologies have helped to improve our understanding of the bacterial communities associated with insects, shedding light on their wide taxonomic and functional diversity. To date, little is known about the microbiota of lepidopterans, which includes some of the most damaging agricultural and forest pests worldwide. Studying their microbiota could help us better understand their ecology and offer insights into developing new pest control strategies. In this paper, we review the literature pertaining to the microbiota of lepidopterans with a focus on pests, and highlight potential recurrent patterns regarding microbiota structure and composition. Keywords: symbiosis; bacterial communities; crop pests; forest pests; Lepidoptera; next generation sequencing (NGS) technologies; diet; developmental stages 1. Introduction Insects represent the most successful taxa of eukaryotic life, being able to colonize almost all environments, including Antarctica, which is populated by some species of chironomids (e.g., Belgica antarctica, Eretmoptera murphyi, and Parochlus steinenii)[1,2]. Many insects are beneficial to plants, playing important roles in seed dispersal, pollination, and plant defense (by feeding upon herbivores, for example) [3]. On the other hand, there are also damaging insects that feed on crops, forest and ornamental plants, or stored products, and, for these reasons, are they considered pests. -

Classical Biological Control of Arthropods in Australia
Classical Biological Contents Control of Arthropods Arthropod index in Australia General index List of targets D.F. Waterhouse D.P.A. Sands CSIRo Entomology Australian Centre for International Agricultural Research Canberra 2001 Back Forward Contents Arthropod index General index List of targets The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its primary mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Australian and developing country researchers in fields where Australia has special competence. Where trade names are used this constitutes neither endorsement of nor discrimination against any product by the Centre. ACIAR MONOGRAPH SERIES This peer-reviewed series contains the results of original research supported by ACIAR, or material deemed relevant to ACIAR’s research objectives. The series is distributed internationally, with an emphasis on the Third World. © Australian Centre for International Agricultural Research, GPO Box 1571, Canberra ACT 2601, Australia Waterhouse, D.F. and Sands, D.P.A. 2001. Classical biological control of arthropods in Australia. ACIAR Monograph No. 77, 560 pages. ISBN 0 642 45709 3 (print) ISBN 0 642 45710 7 (electronic) Published in association with CSIRO Entomology (Canberra) and CSIRO Publishing (Melbourne) Scientific editing by Dr Mary Webb, Arawang Editorial, Canberra Design and typesetting by ClarusDesign, Canberra Printed by Brown Prior Anderson, Melbourne Cover: An ichneumonid parasitoid Megarhyssa nortoni ovipositing on a larva of sirex wood wasp, Sirex noctilio. Back Forward Contents Arthropod index General index Foreword List of targets WHEN THE CSIR Division of Economic Entomology, now Commonwealth Scientific and Industrial Research Organisation (CSIRO) Entomology, was established in 1928, classical biological control was given as one of its core activities. -

Annual Newsletter and Bibliography of the International Society of Plecopterologists PERLA NO. 37, 2019
PERLA Annual Newsletter and Bibliography of The International Society of Plecopterologists Nemoura cinerea (Retzius, 1783) (Nemouridae): Slovenia, near Planina, cave entrance to Ucina River, 15 June 2008. Photograph by Bill P. Stark PERLA NO. 37, 2019 Department of Bioagricultural Sciences and Pest Management Colorado State University Fort Collins, Colorado 80523 USA PERLA Annual Newsletter and Bibliography of the International Society of Plecopterologists Available on Request to the Managing Editor MANAGING EDITOR: Boris C. Kondratieff Department of Bioagricultural Sciences and Pest Management Colorado State University Fort Collins, Colorado 80523 USA E-mail: [email protected] EDITORIAL BOARD: Richard W. Baumann Department of Biology and Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 USA E-mail: [email protected] J. Manuel Tierno de Figueroa Dpto. de Zoología Facultad de Ciencias Universidad de Granada 18071 Granada, SPAIN E-mail: [email protected] Shigekazu Uchida Aichi Institute of Technology 1247 Yagusa Toyota 470-0392, JAPAN E-mail: [email protected] Peter Zwick Schwarzer Stock 9 D-36110 Schlitz, GERMANY E-mail: [email protected] 1 TABLE OF CONTENTS Subscription policy... ............................................................................................................ 3 The XVth International Conference on Ephemeroptera and XIXth International Symposium on Plecoptera ............................................................................................................................. -
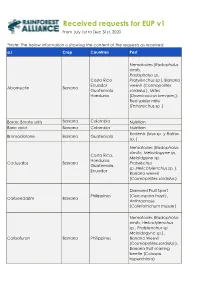
IIS 2020 Requests Overview
Received requests for EUP v1 From July 1st to Dec 31st, 2020 *Note: The below information is showing the content of the requests as received. a.i Crop Countries Pest Nematodes ( Radophulus similis, Radopholus sp, Costa Rica Pratylenchus sp ), Banana Ecuador weevil ( Cosmopolites Abamectin Banana Guatemala sordidus ), Mites Honduras (Dysmicoccus brevipes ), Red spider mite (Tetranychus sp .) Borax; Borate salts Banana Colombia Nutrition Boric acid Banana Colombia Nutrition Rodents ( Mus sp. y Rattus Bromadiolone Banana Guatemala sp. ) Nematodes ( Radopholus silmillis, Meloidogyne sp, Costa Rica, Meloidgune sp, Honduras, Cadusafos Banana Pratylechus Guatemala, sp.,Helicotylenchus sp. ), Ecuador Banana weevil (Cosmopolites sordidus ) Diamond Fruit Sport Philippines (Cercospora hayii ), Carbendazim Banana Anthracnose (Colletotrichum musae ) Nematod es (Radopholus similis, Helicotylenchus sp., Pratylenchus sp. Meloidogyne sp.) , Carbofuran Banana Philippines Banana Weevil (Cosmopolites sordidus ), Banana fruit scarring beetle (Colaspis hyperchlora) a.i Crop Countries Pest Black Sigatoka Ecuador, Costa (Mycosphaerella fijiensis ), Rica, Philippines, Yellow Sigatoka Chlorothalonil Banana Honduras, (Mycosphaerella Guatemala, musicola ), Banana Colombia Freckle ( Phyllosticta musarum ) Mealybug ( Dysmicoccus brevipes, Pseudococcus elisae, Pseudococcus sp, Ferrista sp ), Dysmicoccus sp , Scale insects (Aspidiotus destructor, Diaspis boisduvalii ), Thrips (Thrips florum, Franckliniella spp, Chaetanaphothrips signipennis ), Banana fruit Philippines, -

An Assessment of Biological Control of the Banana Pseudostem Weevil Odoiporus Longicollis (Olivier) by Entomopathogenic Fungi Beauveria Bassiana T
Biocatalysis and Agricultural Biotechnology 20 (2019) 101262 Contents lists available at ScienceDirect Biocatalysis and Agricultural Biotechnology journal homepage: www.elsevier.com/locate/bab An assessment of biological control of the banana pseudostem weevil Odoiporus longicollis (Olivier) by entomopathogenic fungi Beauveria bassiana T Alagersamy Alagesana, Balakrishnan Padmanabanb, Gunasekaran Tharania, Sundaram Jawahara, Subramanian Manivannana,c,* a PG and Research Department of Biotechnology, Bharath College of Science and Management, Thanjavur, 613 005, Tamil Nadu, India b Division of Crop Protection, National Research Centre for Banana (ICAR), Tiruchirappalli, 620 102, Tamil Nadu, India c Department of Zoology, Kongunadu Arts and Science College, Coimbatore, 641 029, Tamil Nadu, India ARTICLE INFO ABSTRACT Keywords: Banana (Musa sp.) is the most imperative staple food crop for all types of people worldwide, which is commonly Banana production grown in Southeast Asia. Banana plantain can be severely affected by the devastating pest Odoiporus longicollis Odoiporus longicollis that results in severe economic losses in India. Management of weevil pests using chemical methods is harmful to Beauveria bassiana the environment, and cultural methods are also partially successful. Therefore, an alternative approach of plant Bioefficacy defense mediated by endophytic fungi to control banana stem borer larvae is necessary, which could affect the Extracellular enzyme extracellular enzyme chitinase and protease. Among four isolates, Beauveria bassiana isolate KH3 is the most Phylogeny virulent entomopathogenic fungus compared with other isolates, and species identification was achieved using molecular phylogenetic characteristics. The B. bassiana isolate KH3 (1 × 108 conidia/mL-1) is more bioeffective against O. longicollis larvae, causing > 90% significant mortality in 12 and 18 days. -

Glossary of Major Terms and Concepts
Glossary of Major Terms and Concepts The following terms and concepts form the core of much of this book. Generally, they appear in boldface type in the text. The chapters in which they are more fully discussed are also given. Acceleration: Faster rate of developmenral evenrs (at any level: cell, organ, individual) in the descendanr; produces peramorphie traits when expressed in the adult pheno type. (Chapters 1 and 2) Allometric heterochrony: Change in a trair as a function of size (as opposed to a function of age in " true" heterochrony); it is useful as adescriptor and can be inrerpretive considering that body size is often a beuer metric of " inrrinsic" age than chronologi cal age. The same categories apply as in " true" heterochrony, only the adjective "allometric" is prefixed: e.g. , " allometric progenesis" is when the descendant ter minates growth at a smaller size (as opposed to age) than the ancestor. (Chapter 2) Allometry: The study of size and shape, usually using biometrie data; the change in size and shape observed. Basic kinds of allometric change include the following. Com plex allometry: occurs when the ratio of the specific growth rates of the traits compared are not constant, a log-log plot comparing the traits will not yield a straight line (i.e., " k" in the allometric formula is inconstanr). Isometry: occurs when there is no change in shape with size increase; that is, when the traits being compared on a log-log plot yield a straight line with a slope (k in the allometric formula) that is effectively equal to 1. -

Table of Contents 2
Southwest Association of Freshwater Invertebrate Taxonomists (SAFIT) List of Freshwater Macroinvertebrate Taxa from California and Adjacent States including Standard Taxonomic Effort Levels 1 March 2011 Austin Brady Richards and D. Christopher Rogers Table of Contents 2 1.0 Introduction 4 1.1 Acknowledgments 5 2.0 Standard Taxonomic Effort 5 2.1 Rules for Developing a Standard Taxonomic Effort Document 5 2.2 Changes from the Previous Version 6 2.3 The SAFIT Standard Taxonomic List 6 3.0 Methods and Materials 7 3.1 Habitat information 7 3.2 Geographic Scope 7 3.3 Abbreviations used in the STE List 8 3.4 Life Stage Terminology 8 4.0 Rare, Threatened and Endangered Species 8 5.0 Literature Cited 9 Appendix I. The SAFIT Standard Taxonomic Effort List 10 Phylum Silicea 11 Phylum Cnidaria 12 Phylum Platyhelminthes 14 Phylum Nemertea 15 Phylum Nemata 16 Phylum Nematomorpha 17 Phylum Entoprocta 18 Phylum Ectoprocta 19 Phylum Mollusca 20 Phylum Annelida 32 Class Hirudinea Class Branchiobdella Class Polychaeta Class Oligochaeta Phylum Arthropoda Subphylum Chelicerata, Subclass Acari 35 Subphylum Crustacea 47 Subphylum Hexapoda Class Collembola 69 Class Insecta Order Ephemeroptera 71 Order Odonata 95 Order Plecoptera 112 Order Hemiptera 126 Order Megaloptera 139 Order Neuroptera 141 Order Trichoptera 143 Order Lepidoptera 165 2 Order Coleoptera 167 Order Diptera 219 3 1.0 Introduction The Southwest Association of Freshwater Invertebrate Taxonomists (SAFIT) is charged through its charter to develop standardized levels for the taxonomic identification of aquatic macroinvertebrates in support of bioassessment. This document defines the standard levels of taxonomic effort (STE) for bioassessment data compatible with the Surface Water Ambient Monitoring Program (SWAMP) bioassessment protocols (Ode, 2007) or similar procedures. -

Downloaded from BOLD Or Requested from Other Authors
www.nature.com/scientificreports OPEN Towards a global DNA barcode reference library for quarantine identifcations of lepidopteran Received: 28 November 2018 Accepted: 5 April 2019 stemborers, with an emphasis on Published: xx xx xxxx sugarcane pests Timothy R. C. Lee 1, Stacey J. Anderson2, Lucy T. T. Tran-Nguyen3, Nader Sallam4, Bruno P. Le Ru5,6, Desmond Conlong7,8, Kevin Powell 9, Andrew Ward10 & Andrew Mitchell1 Lepidopteran stemborers are among the most damaging agricultural pests worldwide, able to reduce crop yields by up to 40%. Sugarcane is the world’s most prolifc crop, and several stemborer species from the families Noctuidae, Tortricidae, Crambidae and Pyralidae attack sugarcane. Australia is currently free of the most damaging stemborers, but biosecurity eforts are hampered by the difculty in morphologically distinguishing stemborer species. Here we assess the utility of DNA barcoding in identifying stemborer pest species. We review the current state of the COI barcode sequence library for sugarcane stemborers, assembling a dataset of 1297 sequences from 64 species. Sequences were from specimens collected and identifed in this study, downloaded from BOLD or requested from other authors. We performed species delimitation analyses to assess species diversity and the efectiveness of barcoding in this group. Seven species exhibited <0.03 K2P interspecifc diversity, indicating that diagnostic barcoding will work well in most of the studied taxa. We identifed 24 instances of identifcation errors in the online database, which has hampered unambiguous stemborer identifcation using barcodes. Instances of very high within-species diversity indicate that nuclear markers (e.g. 18S, 28S) and additional morphological data (genitalia dissection of all lineages) are needed to confrm species boundaries. -

Crambidae Biosecurity Occurrence Background Subfamilies Short Description Diagnosis
Diaphania nitidalis Chilo infuscatellus Crambidae Webworms, Grass Moths, Shoot Borers Biosecurity BIOSECURITY ALERT This Family is of Biosecurity Concern Occurrence This family occurs in Australia. Background The Crambidae is a large, diverse and ubiquitous family of moths that currently comprises 11,500 species globally, with at least half that number again undescribed. The Crambidae and the Pyralidae constitute the superfamily Pyraloidea. Crambid larvae are concealed feeders with a great diversity in feeding habits, shelter building and hosts, such as: leaf rollers, shoot borers, grass borers, leaf webbers, moss feeders, root feeders that shelter in soil tunnels, and solely aquatic life habits. Many species are economically important pests in crops and stored food products. Subfamilies Until recently, the Crambidae was treated as a subfamily under the Pyralidae (snout moths or grass moths). Now they form the superfamily Pyraloidea with the Pyralidae. The Crambidae currently consists of the following 14 subfamilies: Acentropinae Crambinae Cybalomiinae Glaphyriinae Heliothelinae Lathrotelinae Linostinae Midilinae Musotiminae Odontiinae Pyraustinae Schoenobiinae Scopariinae Spilomelinae Short Description Crambid caterpillars are generally cylindrical, with a semiprognathous head and only primary setae (Fig 1). They are often plainly coloured (Fig. 16, Fig. 19), but can be patterned with longitudinal stripes and pinacula that may give them a spotted appearance (Fig. 10, Fig. 11, Fig. 14, Fig. 22). Prolegs may be reduced in borers (Fig. 16). More detailed descriptions are provided below. This factsheet presents, firstly, diagnostic features for the Pyraloidea (Pyralidae and Crambidae) and then the Crambidae. Information and diagnostic features are then provided for crambids listed as priority biosecurity threats for northern Australia. -

Early Shoot Borer on Sugarcane Chilo Infuscatellus Vernacular Name: Illam Kuruthu Puzhu
PEST MANAGEMENT DECISION GUIDE: GREEN AND YELLOW LIST Early Shoot Borer on Sugarcane Chilo infuscatellus Vernacular name: Illam kuruthu Puzhu. Prevention Monitoring Direct Control Direct Control Restrictions l Planting to be done l Monitor for: l Spray Granulosis Virus (750 numbers l Repeated use of same insecticide should be avoided. in early season (Dec l Presence of egg masses under the of diseased larvae) 200ml per acre. l Avoid spraying insecticides up to five to seven days – Jan) surface of the leaves. l Release Trichogramma chilonis 2cc per after parasitoid release. l Avoid moisture l Presence of dead hearts which can acre /release for three times at fifteen stress in the early days interval starting from 30 days after be easily pulled out. l Spray chlorantraniliprole 18.5% l WHO Class U stages of crop, e.g. planting. SC @ 375 ml per ha. Diamide (Unlikely to by making small l Presence of bore holes at the collar l Release 125 gravid females of pesticide. Stomach and contact present acute bundles of plant region. Bore hole at the collar region Sturmiopsis inferens/ha on 30 and 45 poison. hazard in normal (TNAU Agritech Portal) trash and put them in l Light trap catches @ one per five days after planting. use). furrows. hectare. l Spray neem seed kernel extract 5%. l Intercropping with l Monitoring through pheromone (25 kg of NSK/ha dissolved in 500 litres l Spray fipronil 5% SC @ 2 litres l WHO Class II dhaincha or pulses to traps @ 5 per acre. Erect the trap of water). per ha. -

Management of Insect Pests by Microorganisms
Proc Indian Natn Sci Acad 80 No. 2 June 2014 Spl. Sec. pp. 455-471 Printed in India. Review Article Management of Insect Pests by Microorganisms B RAMANUJAM*, R RANGESHWARAN, G SIVAKMAR, M MOHAN and M S YANDIGERI National Bureau of Agriculturally Important Insects, P.B. No. 2491, HA Farm Post, Bellary Road, Bangalore 560024, Karnataka, India (Received on 09 April 2013; Revised on 20 August 2013; Accepted on 23 September 2013) Insects, like other organisms, are susceptible to a variety of diseases caused by bacteria, viruses, fungi and protozoans, and these pathogens are exploited for biological control of insect pests through introductory or inundative applications. Microbial pathogens of insects are intensively investigated to develop environmental friendly pest management strategies in agriculture and forestry. In this paper, the scope for utilization of insect pathogens in pest management in the world and India is reviewed. The most successfully utilized insect pathogen is the bacterium, Bacillus thuringiensis (Bt) which is used extensively for management of certain lepidopteran pests. In India, mostly imported products of Bt kurstaki have been used, which are expensive and there is an urgent need to develop aggressive indigenous Bt strains against various pests. Baculoviruses comprising nuclear polyhedrosis virus (NPV) and granulosis virus (GV) have been successfully used as insect pathogens because of their high virulence and specificity. NPV and GV formulations are used for lepidopteran pests like Helicoverpa armigera (HaNPV) and Spodoptera litura (SlNPV) in India, besides Anticarsia gemmatalis NPV in Brazil, Lymanttria disper NPV, Orgyia pseudotsugata NPV in USA and GV of Pieris rapae in China. Lack of easy mass multiplication methods for the commercial production of baculoviruses calls for R&D to develop production in insect tissue cultures. -

2019 Cover Art (In Collaboration with 4-H Youth Development, University of Arizona Cooperative Extension)
Association for the Advancement of Industrial Crops Advancing the adoption of industrial crops through innovation and technology El Conquistador Hilton Resort Tucson, Arizona USA September 8-11, 2019 Cover art (in collaboration with 4-H Youth Development, University of Arizona Cooperative Extension) PLANT MATTER MAKES THE WORLD GO ROUND Alexis Peck, Grade 11, Duncan High School, Duncan, AZ ASSOCIATION FOR THE ADVANCEMENT OF INDUSTRIAL CROPS www.aaic.org “ADVANCING THE ADOPTION OF INDUSTRIAL CROPS THROUGH INNOVATION AND TECHNOLOGY” 31st Annual Meeting September 8-11, 2019 Tucson, Arizona USA Sponsors i AAIC BOARD OF DIRECTORS President: Von Mark V. Cruz, Bridgestone Americas, Inc., Eloy, AZ, USA President-Elect: Federica Zanetti, University of Bologna, Bologna, Italy Past-President: Jim Todd, Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA), Simcoe, ON, Canada Secretary: Claire C. Heinitz, USDA-ARS National Arid Land Plant Genetic Resources Unit, Parlier, CA, USA Treasurer: Burton L. Johnson, North Dakota State University, Fargo, ND, USA Registrar / Valerie H. Teetor, University of Arizona, Tucson, AZ, USA Membership: Webmaster: Von Mark V. Cruz, Bridgestone Americas, Inc., Eloy, AZ, USA Division Chairs Fibers & Cellulosics: Efthymia Alexopoulou, Centre for Renewable Energy Sources and Saving (CRES), Pikermi, Greece General Crops & Ana Luisa Fernando, Nova University of Lisbon, Lisbon, Portugal Products: Medicinal & Diana Jasso De Rodríguez, Universidad Autónoma Agraria Antonio Nutraceutical Plants: Narro, Saltillo, Coahuila, México Natural Rubber & Guangyao (Sam) Wang, Bridgestone Americas, Inc., Eloy, AZ, USA Resins: Oilseeds: Hussein Abdel-Haleem, USDA-ARS ALARC, Maricopa, AZ, USA To cite this publication: Cruz, V.M.V. and M. Berti. (eds.) 2019. Advancing the Adoption of Industrial Crops through Innovation and Technology.