ZNF44 (NM 016264) Human Tagged ORF Clone Product Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Abbott Molecular Oncology and Genetics 2016 U.S
DESCRIPTOR, 9/12, ALL CAPS ABBOTT MOLECULAR ONCOLOGY AND GENETICS 2016 U.S. Product Catalog Area for placed imagery Only use imagery that is relevant to the communication CHOOSE TRANSFORMATION See where it will take you at AbbottMolecular.com 2 ASR Analyte Specific Reagent GPR General Purpose Reagent IVD In Vitro Diagnostic RUO Research Use Only All products manufactured and/or distributed by Abbott Molecular should be used in accordance with the products’ labeled intended use. Products labeled “Research Use Only” should be used for research applications, and are not for use in diagnostic procedures. CEP, LSI, AneuVysion, MultiVysion, PathVysion and Vysis are registered trademarks of Vysis, Inc., AutoVysion, ProbeChek, SpectrumAqua, SpectrumBlue, SpectrumGreen, SpectrumGold, SpectrumOrange, SpectrumRed, SpectrumFRed, TelVysion, ToTelVysion, UroVysion and VP 2000 are trademarks of Abbott Molecular in various jurisdictions. All other trademarks are the property of their respective owners. 3 Abbott Molecular is Transforming Laboratory Partnerships and Productivity—Today and into the Future As a leader in molecular diagnostics, Our commitment to exploring new clinical frontiers is evident in the development and delivery of innovative systems and Abbott is committed to providing assay solutions that aid physicians in the diagnosis of disease, selection of therapies and monitoring of disease. solution-oriented oferings built The new product oferings in this catalog and those coming on FISH and PCR. Building on throughout the remainder of 2016 have been designed in partnership with laboratories, directly incorporating the a proven track record of service feedback we’ve gathered from you. These options expand the Vysis FISH portfolio and increase productivity by to the worldwide community of driving improvements in laboratory efciency and enabling researchers and clinicians, Abbott customization of solutions on a lab by lab basis. -
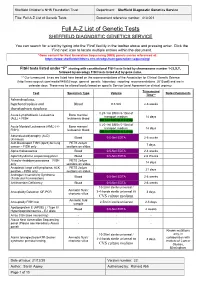
Full A-Z List of Genetic Tests Document Reference Number: 413.001
Sheffield Children’s NHS Foundation Trust Department: Sheffield Diagnostic Genetics Service Title: Full A-Z List of Genetic Tests Document reference number: 413.001 Full A-Z List of Genetic Tests SHEFFIELD DIAGNOSTIC GENETICS SERVICE You can search for a test by typing into the ‘Find’ facility in the toolbar above and pressing enter. Click the ‘Find next’ icon to locate multiple entries within the document. *Gene content for Next Generation Sequencing (NGS) panels can be referenced at: https://www.sheffieldchildrens.nhs.uk/sdgs/next-generation-sequencing/ FISH tests listed under “F” starting with constitutional FISH tests listed by chromosome number 1-22,X,Y, followed by oncology FISH tests listed A-Z by gene name. ** Our turnaround times are listed here based on the recommendations of the Association for Clinical Genetic Science (http://www.acgs.uk.com/media/949852/acgs_general_genetic_laboratory_reporting_recommendations_2015.pdf) and are in calendar days. These may be altered locally based on specific Service Level Agreement or clinical urgency. Turnaround Test Specimen Type Volume Notes/Comments Time** Achondroplasia, hypchondroplasia and Blood 0.5-5ml 2-6 weeks thanatophoric dysplasia 0.25-1ml BM in 5-10ml of Acute Lymphoblastic Leukaemia Bone marrow/ transport medium 14 days (ALL) + FISH leukaemic blood OR 1ml BM/VB in Li Hep 0.25-1ml BM in 5-10ml of Acute Myeloid Leukaemia (AML) (+/- Bone marrow/ transport medium 14 days FISH) leukaemic blood OR 1ml BM/VB in Li Hep Adrenoleukodystrophy (ALD) Blood 0.5-5ml EDTA 2-6 weeks (X-linked) -

Supplementary Table 3 Gene Microarray Analysis: PRL+E2 Vs
Supplementary Table 3 Gene microarray analysis: PRL+E2 vs. control ID1 Field1 ID Symbol Name M Fold P Value 69 15562 206115_at EGR3 early growth response 3 2,36 5,13 4,51E-06 56 41486 232231_at RUNX2 runt-related transcription factor 2 2,01 4,02 6,78E-07 41 36660 227404_s_at EGR1 early growth response 1 1,99 3,97 2,20E-04 396 54249 36711_at MAFF v-maf musculoaponeurotic fibrosarcoma oncogene homolog F 1,92 3,79 7,54E-04 (avian) 42 13670 204222_s_at GLIPR1 GLI pathogenesis-related 1 (glioma) 1,91 3,76 2,20E-04 65 11080 201631_s_at IER3 immediate early response 3 1,81 3,50 3,50E-06 101 36952 227697_at SOCS3 suppressor of cytokine signaling 3 1,76 3,38 4,71E-05 16 15514 206067_s_at WT1 Wilms tumor 1 1,74 3,34 1,87E-04 171 47873 238623_at NA NA 1,72 3,30 1,10E-04 600 14687 205239_at AREG amphiregulin (schwannoma-derived growth factor) 1,71 3,26 1,51E-03 256 36997 227742_at CLIC6 chloride intracellular channel 6 1,69 3,23 3,52E-04 14 15038 205590_at RASGRP1 RAS guanyl releasing protein 1 (calcium and DAG-regulated) 1,68 3,20 1,87E-04 55 33237 223961_s_at CISH cytokine inducible SH2-containing protein 1,67 3,19 6,49E-07 78 32152 222872_x_at OBFC2A oligonucleotide/oligosaccharide-binding fold containing 2A 1,66 3,15 1,23E-05 1969 32201 222921_s_at HEY2 hairy/enhancer-of-split related with YRPW motif 2 1,64 3,12 1,78E-02 122 13463 204015_s_at DUSP4 dual specificity phosphatase 4 1,61 3,06 5,97E-05 173 36466 227210_at NA NA 1,60 3,04 1,10E-04 117 40525 231270_at CA13 carbonic anhydrase XIII 1,59 3,02 5,62E-05 81 42339 233085_s_at OBFC2A oligonucleotide/oligosaccharide-binding -

Single Cell Transcriptomics in Schizophrenia Postmortem Brain: Moving Beyond Bulk Lysate
SINGLE CELL TRANSCRIPTOMICS IN SCHIZOPHRENIA POSTMORTEM BRAIN: MOVING BEYOND BULK LYSATE Richard Crist June 8th, 2020 Schizophrenia Transcriptomics ■ Microarray and RNA sequencing ■ Differentially expressed genes across many cortical and sub-cortical regions – Dorsolateral prefrontal cortex (dlPFC) (Fillman et al, 2013) – Anterior Cingulate Cortex (Zhao et al, 2015; Hong et al, 2013) – Superior temporal gyrus (Wu et al, 2012) – Hippocampus (Hwang et al, 2013; Kohen et al, 2014) – Amygdala (Chang et al, 2017) ■ Enrichment of pathways and gene networks – Neural development – Axon guidance – Inflammation and immune-related proteins CommonMind Consortium ■ Largest transcriptomic analysis of schizophrenia – 258 cases/279 controls – RNAseq in dlPFC ■ 693 differentially expressed genes Fromer et al, 2016 Cell Diversity in Postmortem Brain ■ Brain, like all tissues, consists of many cell types – Major cell populations (e.g. astrocytes) – Distinct sub-populations (e.g. PVALB+ interneurons) ■ Problems in assessing differential expression in bulk lysate – Inability to identify which cells are affected – Missed expression changes in less common cell types Penney et al, 2019 Schizophrenia Single Cell Transcriptomics ■ Immunofluorescence and laser capture microdissection to collect individual populations of cells ■ Layer III/V pyramidal neurons (Arion et al, 2017) – 72 PFC samples – 36 cases/36 controls – 100 cells per layer for each sample – Expression assessed by microarray – 1,783 differentially expressed probe sets corresponding to 1,420 genes -

Targeted Sequence Capture and Ultra High Throughput Sequencing for Gene Discovery in Inherited Diseases
Unicentre CH-1015 Lausanne http://serval.unil.ch Year : 2013 Targeted sequence capture and Ultra High Throughput sequencing for gene discovery in inherited diseases Dl GIOIA SILVIO ALESSANDRO Dl GIOIA SILVIO ALESSANDRO, 2013, Targeted sequence capture and Ultra High Throughput sequencing for gene discovery in inherited diseases Originally published at : Thesis, University of Lausanne Posted at the University of Lausanne Open Archive. http://serval.unil.ch Droits d’auteur L'Université de Lausanne attire expressément l'attention des utilisateurs sur le fait que tous les documents publiés dans l'Archive SERVAL sont protégés par le droit d'auteur, conformément à la loi fédérale sur le droit d'auteur et les droits voisins (LDA). A ce titre, il est indispensable d'obtenir le consentement préalable de l'auteur et/ou de l’éditeur avant toute utilisation d'une oeuvre ou d'une partie d'une oeuvre ne relevant pas d'une utilisation à des fins personnelles au sens de la LDA (art. 19, al. 1 lettre a). A défaut, tout contrevenant s'expose aux sanctions prévues par cette loi. Nous déclinons toute responsabilité en la matière. Copyright The University of Lausanne expressly draws the attention of users to the fact that all documents published in the SERVAL Archive are protected by copyright in accordance with federal law on copyright and similar rights (LDA). Accordingly it is indispensable to obtain prior consent from the author and/or publisher before any use of a work or part of a work for purposes other than personal use within the meaning of LDA (art. 19, para. -

The Effect of Compound L19 on Human Colorectal Cells (DLD-1)
Stephen F. Austin State University SFA ScholarWorks Electronic Theses and Dissertations Spring 5-16-2018 The Effect of Compound L19 on Human Colorectal Cells (DLD-1) Sepideh Mohammadhosseinpour [email protected] Follow this and additional works at: https://scholarworks.sfasu.edu/etds Part of the Biotechnology Commons Tell us how this article helped you. Repository Citation Mohammadhosseinpour, Sepideh, "The Effect of Compound L19 on Human Colorectal Cells (DLD-1)" (2018). Electronic Theses and Dissertations. 188. https://scholarworks.sfasu.edu/etds/188 This Thesis is brought to you for free and open access by SFA ScholarWorks. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of SFA ScholarWorks. For more information, please contact [email protected]. The Effect of Compound L19 on Human Colorectal Cells (DLD-1) Creative Commons License This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 4.0 License. This thesis is available at SFA ScholarWorks: https://scholarworks.sfasu.edu/etds/188 The Effect of Compound L19 on Human Colorectal Cells (DLD-1) By Sepideh Mohammadhosseinpour, Master of Science Presented to the Faculty of the Graduate School of Stephen F. Austin State University In Partial Fulfillment Of the Requirements For the Degree of Master of Science in Biotechnology STEPHEN F. AUSTIN STATE UNIVERSITY May, 2018 The Effect of Compound L19 on Human Colorectal Cells (DLD-1) By Sepideh Mohammadhosseinpour, Master of Science APPROVED: Dr. Beatrice A. Clack, Thesis Director Dr. Josephine Taylor, Committee Member Dr. Rebecca Parr, Committee Member Dr. Stephen Mullin, Committee Member Pauline Sampson, Ph.D. -

WO 2012/054896 Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number ι (43) International Publication Date ¾ ί t 2 6 April 2012 (26.04.2012) WO 2012/054896 Al (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, C12N 5/00 (2006.01) C12N 15/00 (2006.01) CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, C12N 5/02 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (21) International Application Number: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, PCT/US201 1/057387 ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, (22) International Filing Date: NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, 2 1 October 201 1 (21 .10.201 1) RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, (25) Filing Language: English ZM, ZW. (26) Publication Language: English (84) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of regional protection available): ARIPO (BW, GH, 61/406,064 22 October 2010 (22.10.2010) US GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, 61/415,244 18 November 2010 (18.1 1.2010) US UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, (71) Applicant (for all designated States except US): BIO- DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, TIME INC. -

Transforming Growth Factor ß1-Mediated Functional Inhibition Of
Myelodysplastic Syndromes SUPPLEMENTARY APPENDIX Transforming growth factor 1- mediated functional inhibition of mesenchymal stromal celβls in myelodysplastic syndromes and acute myeloid leukemia Stefanie Geyh, 1* Manuel Rodríguez-Paredes, 1,2 * Paul Jäger, 1 Annemarie Koch, 1 Felix Bormann, 2 Julian Gutekunst, 2 Christoph Zilkens, 3 Ulrich Germing, 1 Guido Kobbe, 1 Frank Lyko, 2 Rainer Haas 1 and Thomas Schroeder 1 1Department of Hematology, Oncology and Clinical Immunology, University of Duesseldorf, Medical Faculty; 2Division of Epigenetics, DKFZ- ZMBH Alliance, German Cancer Research Center, Heidelberg and 3Department of Orthopedic Surgery, University of Duesseldorf, Medical Faculty, Germany *SG and MR-P contributed equally to this work. ©2018 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.186734 Received: December 19, 2017. Accepted: May 14, 2018. Pre-published: May 17, 2018. Correspondence: [email protected] Figure S1 Downregulated genes Downregulated genes Upregulated Figure S1. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 9 RCMD-, RAEB- and AML-derived MSC samples. Color scale depicts the rlog-transformed FPKM values for each gene and every sample. Figure S2 Downregulated genes Downregulated genes Upregulated Figure S2. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 3 RCMD, RAEB and AML MSC samples, respectively. Color scales depict the rlog-transformed FPKM values for each gene and every sample. Figure S3 A. B. 0.0015 *** ** <-3 -2 0.0010 RCMD RAEB AML -1 0 1 0.0005 Log2FC LTF 2 CCL26/GAPDH INHBB >3 0.0000 TGFB2 y S h D ML M A ealt ll LTF H a EGF 0.003 *** ** INHBB TGFB2 0.002 INHBB IGFBP7 0.001 GDF11 LIF/GAPDH BMP1 0.000 y L th M TNFSF12 l A FGF13 ea ll MDS H a FGF13 0.0015 * TNFSF10 TNFSF10 0.0010 0.0005 SPP1/GAPDH 0.0000 y th l AML ea H all MDS Figure S3. -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization. -

Control of the Physical and Antimicrobial Skin Barrier by an IL-31–IL-1 Signaling Network
The Journal of Immunology Control of the Physical and Antimicrobial Skin Barrier by an IL-31–IL-1 Signaling Network Kai H. Ha¨nel,*,†,1,2 Carolina M. Pfaff,*,†,1 Christian Cornelissen,*,†,3 Philipp M. Amann,*,4 Yvonne Marquardt,* Katharina Czaja,* Arianna Kim,‡ Bernhard Luscher,€ †,5 and Jens M. Baron*,5 Atopic dermatitis, a chronic inflammatory skin disease with increasing prevalence, is closely associated with skin barrier defects. A cy- tokine related to disease severity and inhibition of keratinocyte differentiation is IL-31. To identify its molecular targets, IL-31–dependent gene expression was determined in three-dimensional organotypic skin models. IL-31–regulated genes are involved in the formation of an intact physical skin barrier. Many of these genes were poorly induced during differentiation as a consequence of IL-31 treatment, resulting in increased penetrability to allergens and irritants. Furthermore, studies employing cell-sorted skin equivalents in SCID/NOD mice demonstrated enhanced transepidermal water loss following s.c. administration of IL-31. We identified the IL-1 cytokine network as a downstream effector of IL-31 signaling. Anakinra, an IL-1R antagonist, blocked the IL-31 effects on skin differentiation. In addition to the effects on the physical barrier, IL-31 stimulated the expression of antimicrobial peptides, thereby inhibiting bacterial growth on the three-dimensional organotypic skin models. This was evident already at low doses of IL-31, insufficient to interfere with the physical barrier. Together, these findings demonstrate that IL-31 affects keratinocyte differentiation in multiple ways and that the IL-1 cytokine network is a major downstream effector of IL-31 signaling in deregulating the physical skin barrier.