( 12 ) United States Patent
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

ATRX Induction by Mutant Huntingtin Via Cdx2 Modulates Heterochromatin Condensation and Pathology in Huntington’S Disease
Cell Death and Differentiation (2012) 19, 1109–1116 & 2012 Macmillan Publishers Limited All rights reserved 1350-9047/12 www.nature.com/cdd ATRX induction by mutant huntingtin via Cdx2 modulates heterochromatin condensation and pathology in Huntington’s disease J Lee1,2, YK Hong3, GS Jeon4, YJ Hwang4, KY Kim4, KH Seong4, M-K Jung4, DJ Picketts5, NW Kowall1,2, KS Cho3 and H Ryu*,1,2,4 Aberrant chromatin remodeling is involved in the pathogenesis of Huntington’s disease (HD) but the mechanism is not known. Herein, we report that mutant huntingtin (mtHtt) induces the transcription of alpha thalassemia/mental retardation X linked (ATRX), an ATPase/helicase and SWI/SNF-like chromatin remodeling protein via Cdx-2 activation. ATRX expression was elevated in both a cell line model and transgenic model of HD, and Cdx-2 occupancy of the ATRX promoter was increased in HD. Induction of ATRX expanded the size of promyelocytic leukemia nuclear body (PML-NB) and increased trimethylation of H3K9 (H3K9me3) and condensation of pericentromeric heterochromatin, while knockdown of ATRX decreased PML-NB and H3K9me3 levels. Knockdown of ATRX/dXNP improved the hatch rate of fly embryos expressing mtHtt (Q127). ATRX/dXNP overexpression exacerbated eye degeneration of eye-specific mtHtt (Q127) expressing flies. Our findings suggest that transcriptional alteration of ATRX by mtHtt is involved in pericentromeric heterochromatin condensation and contributes to the pathogenesis of HD. Cell Death and Differentiation (2012) 19, 1109–1116; doi:10.1038/cdd.2011.196; published -
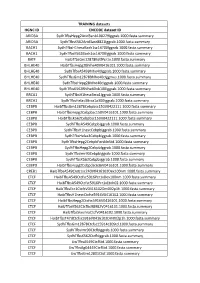
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

Roles of the CSE1L-Mediated Nuclear Import Pathway in Epigenetic
Roles of the CSE1L-mediated nuclear import pathway PNAS PLUS in epigenetic silencing Qiang Donga,b,c, Xiang Lia,b,c, Cheng-Zhi Wangb, Shaohua Xuc, Gang Yuanc, Wei Shaoc, Baodong Liud, Yong Zhengb, Hailin Wangd, Xiaoguang Leic,e,f, Zhuqiang Zhangb,1, and Bing Zhua,b,g,1 aGraduate Program, Peking Union Medical College and Chinese Academy of Medical Sciences, 100730 Beijing, China; bNational Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101 Beijing, China; cNational Institute of Biological Sciences, 102206 Beijing, China; dThe State Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, 100085 Beijing, China; eBeijing National Laboratory for Molecular Sciences, Department of Chemical Biology, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China; fPeking-Tsinghua Center for Life Sciences, Peking University, 100871 Beijing, China; and gCollege of Life Sciences, University of Chinese Academy of Sciences, 100049 Beijing, China Edited by Arthur D. Riggs, Beckman Research Institute of City of Hope, Duarte, CA, and approved March 21, 2018 (received for review January 17, 2018) Epigenetic silencing can be mediated by various mechanisms, CSE1L, a key player in the nuclear import pathway, as an es- and many regulators remain to be identified. Here, we report a sential factor for maintaining the repression of many methyl- genome-wide siRNA screening to identify regulators essential for ated genes. Mechanistically, CSE1L functions by facilitating maintaining gene repression of a CMV promoter silenced by DNA the nuclear import of certain cargo proteins that are essential methylation. -

2017.08.28 Anne Barry-Reidy Thesis Final.Pdf
REGULATION OF BOVINE β-DEFENSIN EXPRESSION THIS THESIS IS SUBMITTED TO THE UNIVERSITY OF DUBLIN FOR THE DEGREE OF DOCTOR OF PHILOSOPHY 2017 ANNE BARRY-REIDY SCHOOL OF BIOCHEMISTRY & IMMUNOLOGY TRINITY COLLEGE DUBLIN SUPERVISORS: PROF. CLIONA O’FARRELLY & DR. KIERAN MEADE TABLE OF CONTENTS DECLARATION ................................................................................................................................. vii ACKNOWLEDGEMENTS ................................................................................................................... viii ABBREVIATIONS ................................................................................................................................ix LIST OF FIGURES............................................................................................................................. xiii LIST OF TABLES .............................................................................................................................. xvii ABSTRACT ........................................................................................................................................xix Chapter 1 Introduction ........................................................................................................ 1 1.1 Antimicrobial/Host-defence peptides ..................................................................... 1 1.2 Defensins................................................................................................................. 1 1.3 β-defensins ............................................................................................................. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

A Clinicopathological and Molecular Genetic Analysis of Low-Grade Glioma in Adults
A CLINICOPATHOLOGICAL AND MOLECULAR GENETIC ANALYSIS OF LOW-GRADE GLIOMA IN ADULTS Presented by ANUSHREE SINGH MSc A thesis submitted in partial fulfilment of the requirements of the University of Wolverhampton for the degree of Doctor of Philosophy Brain Tumour Research Centre Research Institute in Healthcare Sciences Faculty of Science and Engineering University of Wolverhampton November 2014 i DECLARATION This work or any part thereof has not previously been presented in any form to the University or to any other body whether for the purposes of assessment, publication or for any other purpose (unless otherwise indicated). Save for any express acknowledgments, references and/or bibliographies cited in the work, I confirm that the intellectual content of the work is the result of my own efforts and of no other person. The right of Anushree Singh to be identified as author of this work is asserted in accordance with ss.77 and 78 of the Copyright, Designs and Patents Act 1988. At this date copyright is owned by the author. Signature: Anushree Date: 30th November 2014 ii ABSTRACT The aim of the study was to identify molecular markers that can determine progression of low grade glioma. This was done using various approaches such as IDH1 and IDH2 mutation analysis, MGMT methylation analysis, copy number analysis using array comparative genomic hybridisation and identification of differentially expressed miRNAs using miRNA microarray analysis. IDH1 mutation was present at a frequency of 71% in low grade glioma and was identified as an independent marker for improved OS in a multivariate analysis, which confirms the previous findings in low grade glioma studies. -

Farnesol-Induced Apoptosis in Human Lung Carcinoma Cells Is Coupled to the Endoplasmic Reticulum Stress Response
Research Article Farnesol-Induced Apoptosis in Human Lung Carcinoma Cells Is Coupled to the Endoplasmic Reticulum Stress Response Joung Hyuck Joo,1 Grace Liao,1 Jennifer B. Collins,2 Sherry F. Grissom,2 and Anton M. Jetten1 1Cell Biology Section, LRB, and 2Microarray Group, Division of Intramural Research, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, North Carolina Abstract range of fruits and vegetables (9, 10). Each isoprenoid has been Farnesol (FOH) and other isoprenoid alcohols induce apopto- shown to inhibit proliferation and induce apoptosis in a number of sis in various carcinoma cells and inhibit tumorigenesis in neoplastic cell lines from different origins (4, 11–14). In addition, in vivo these isoprenoids have been reported to be effective in chemo- several models. However, the mechanisms by which in vivo they mediate their effects are not yet fully understood. In this prevention and chemotherapy in various cancer models study, we show that FOH is an effective inducer of apoptosis in (10, 12, 15, 16). FOH has been reported to exhibit chemopreventive several lung carcinoma cells, including H460. This induction is effects in colon and pancreas carcinogenesis in rats (9, 17) whereas associated with activation of several caspases and cleavage of phase I and II clinical trials have indicated therapeutic potential poly(ADP-ribose) polymerase (PARP). To obtain insight into for POH (16, 18). The mechanisms by which these isoprenoids induce these effects are not yet fully understood. Isoprenoids have the mechanism involved in FOH-induced apoptosis, we compared the gene expression profiles of FOH-treated and been reported to inhibit posttranslational protein prenylation (19) control H460 cells by microarray analysis. -

Identification of BBX Proteins As Rate-Limiting Co-Factors of HY5
1 Identification of BBX proteins as rate-limiting co-factors of HY5. 2 Katharina Bursch1, Gabriela Toledo-Ortiz2, Marie Pireyre3, Miriam Lohr1, Cordula Braatz1, Henrik 3 Johansson1* 4 1. Institute of Biology/Applied Genetics, Dahlem Centre of Plant Sciences (DCPS), Freie Universität 5 Berlin, Albrecht-Thaer-Weg 6. D-14195 Berlin, Germany. 6 2. Lancaster Environment Centre, Lancaster University, Lancaster LA1 4YQ, UK 7 3. Department of Botany and Plant Biology, Section of Biology, Faculty of Sciences, University of 8 Geneva, 30 Quai E. Ansermet, 1211 Geneva 4, Switzerland 9 *E-mail [email protected] 10 Abstract 11 As a source of both energy and environmental information, monitoring the incoming light is crucial for 12 plants to optimize growth throughout development1. Concordantly, the light signalling pathways in 13 plants are highly integrated with numerous other regulatory pathways2,3. One of these signal 14 integrators is the bZIP transcription factor HY5 which holds a key role as a positive regulator of light 15 signalling in plants4,5. Although HY5 is thought to act as a DNA-binding transcriptional regulator6,7, the 16 lack of any apparent transactivation domain8 makes it unclear how HY5 is able to accomplish its many 17 functions. Here, we describe the identification of three B-box containing proteins (BBX20, 21 and 22) 18 as essential partners for HY5 dependent modulation of hypocotyl elongation, anthocyanin 19 accumulation and transcriptional regulation. The bbx202122 triple mutant mimics the phenotypes of 20 hy5 in the light and its ability to suppress the cop1 mutant phenotype in darkness. Furthermore, 84% 21 of genes that exhibit differential expression in bbx202122 are also HY5 regulated, and we provide 22 evidence that HY5 requires the B-box proteins for transcriptional regulation. -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Epigenetics Page 1
Epigenetics esiRNA ID Gene Name Gene Description Ensembl ID HU-13237-1 ACTL6A actin-like 6A ENSG00000136518 HU-13925-1 ACTL6B actin-like 6B ENSG00000077080 HU-14457-1 ACTR1A ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) ENSG00000138107 HU-10579-1 ACTR2 ARP2 actin-related protein 2 homolog (yeast) ENSG00000138071 HU-10837-1 ACTR3 ARP3 actin-related protein 3 homolog (yeast) ENSG00000115091 HU-09776-1 ACTR5 ARP5 actin-related protein 5 homolog (yeast) ENSG00000101442 HU-00773-1 ACTR6 ARP6 actin-related protein 6 homolog (yeast) ENSG00000075089 HU-07176-1 ACTR8 ARP8 actin-related protein 8 homolog (yeast) ENSG00000113812 HU-09411-1 AHCTF1 AT hook containing transcription factor 1 ENSG00000153207 HU-15150-1 AIRE autoimmune regulator ENSG00000160224 HU-12332-1 AKAP1 A kinase (PRKA) anchor protein 1 ENSG00000121057 HU-04065-1 ALG13 asparagine-linked glycosylation 13 homolog (S. cerevisiae) ENSG00000101901 HU-13552-1 ALKBH1 alkB, alkylation repair homolog 1 (E. coli) ENSG00000100601 HU-06662-1 ARID1A AT rich interactive domain 1A (SWI-like) ENSG00000117713 HU-12790-1 ARID1B AT rich interactive domain 1B (SWI1-like) ENSG00000049618 HU-09415-1 ARID2 AT rich interactive domain 2 (ARID, RFX-like) ENSG00000189079 HU-03890-1 ARID3A AT rich interactive domain 3A (BRIGHT-like) ENSG00000116017 HU-14677-1 ARID3B AT rich interactive domain 3B (BRIGHT-like) ENSG00000179361 HU-14203-1 ARID3C AT rich interactive domain 3C (BRIGHT-like) ENSG00000205143 HU-09104-1 ARID4A AT rich interactive domain 4A (RBP1-like) ENSG00000032219 HU-12512-1 ARID4B AT rich interactive domain 4B (RBP1-like) ENSG00000054267 HU-12520-1 ARID5A AT rich interactive domain 5A (MRF1-like) ENSG00000196843 HU-06595-1 ARID5B AT rich interactive domain 5B (MRF1-like) ENSG00000150347 HU-00556-1 ASF1A ASF1 anti-silencing function 1 homolog A (S.