Variance Components + Random Effects
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lecture 22: Bivariate Normal Distribution Distribution
6.5 Conditional Distributions General Bivariate Normal Let Z1; Z2 ∼ N (0; 1), which we will use to build a general bivariate normal Lecture 22: Bivariate Normal Distribution distribution. 1 1 2 2 f (z1; z2) = exp − (z1 + z2 ) Statistics 104 2π 2 We want to transform these unit normal distributions to have the follow Colin Rundel arbitrary parameters: µX ; µY ; σX ; σY ; ρ April 11, 2012 X = σX Z1 + µX p 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 1 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - Marginals General Bivariate Normal - Cov/Corr First, lets examine the marginal distributions of X and Y , Second, we can find Cov(X ; Y ) and ρ(X ; Y ) Cov(X ; Y ) = E [(X − E(X ))(Y − E(Y ))] X = σX Z1 + µX h p i = E (σ Z + µ − µ )(σ [ρZ + 1 − ρ2Z ] + µ − µ ) = σX N (0; 1) + µX X 1 X X Y 1 2 Y Y 2 h p 2 i = N (µX ; σX ) = E (σX Z1)(σY [ρZ1 + 1 − ρ Z2]) h 2 p 2 i = σX σY E ρZ1 + 1 − ρ Z1Z2 p 2 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY = σX σY ρE[Z1 ] p 2 = σX σY ρ = σY [ρN (0; 1) + 1 − ρ N (0; 1)] + µY = σ [N (0; ρ2) + N (0; 1 − ρ2)] + µ Y Y Cov(X ; Y ) ρ(X ; Y ) = = ρ = σY N (0; 1) + µY σX σY 2 = N (µY ; σY ) Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 2 / 22 Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 3 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - RNG Multivariate Change of Variables Consequently, if we want to generate a Bivariate Normal random variable Let X1;:::; Xn have a continuous joint distribution with pdf f defined of S. -
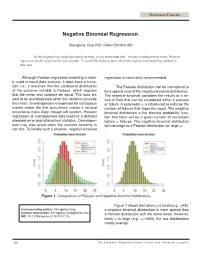
Negative Binomial Regression
STATISTICS COLUMN Negative Binomial Regression Shengping Yang PhD ,Gilbert Berdine MD In the hospital stay study discussed recently, it was mentioned that “in case overdispersion exists, Poisson regression model might not be appropriate.” I would like to know more about the appropriate modeling method in that case. Although Poisson regression modeling is wide- regression is commonly recommended. ly used in count data analysis, it does have a limita- tion, i.e., it assumes that the conditional distribution The Poisson distribution can be considered to of the outcome variable is Poisson, which requires be a special case of the negative binomial distribution. that the mean and variance be equal. The data are The negative binomial considers the results of a se- said to be overdispersed when the variance exceeds ries of trials that can be considered either a success the mean. Overdispersion is expected for contagious or failure. A parameter ψ is introduced to indicate the events where the first occurrence makes a second number of failures that stops the count. The negative occurrence more likely, though still random. Poisson binomial distribution is the discrete probability func- regression of overdispersed data leads to a deflated tion that there will be a given number of successes standard error and inflated test statistics. Overdisper- before ψ failures. The negative binomial distribution sion may also result when the success outcome is will converge to a Poisson distribution for large ψ. not rare. To handle such a situation, negative binomial 0 5 10 15 20 25 0 5 10 15 20 25 Figure 1. Comparison of Poisson and negative binomial distributions. -

Section 1: Regression Review
Section 1: Regression review Yotam Shem-Tov Fall 2015 Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 1 / 58 Contact information Yotam Shem-Tov, PhD student in economics. E-mail: [email protected]. Office: Evans hall room 650 Office hours: To be announced - any preferences or constraints? Feel free to e-mail me. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 2 / 58 R resources - Prior knowledge in R is assumed In the link here is an excellent introduction to statistics in R . There is a free online course in coursera on programming in R. Here is a link to the course. An excellent book for implementing econometric models and method in R is Applied Econometrics with R. This is my favourite for starting to learn R for someone who has some background in statistic methods in the social science. The book Regression Models for Data Science in R has a nice online version. It covers the implementation of regression models using R . A great link to R resources and other cool programming and econometric tools is here. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 3 / 58 Outline There are two general approaches to regression 1 Regression as a model: a data generating process (DGP) 2 Regression as an algorithm (i.e., an estimator without assuming an underlying structural model). Overview of this slides: 1 The conditional expectation function - a motivation for using OLS. 2 OLS as the minimum mean squared error linear approximation (projections). 3 Regression as a structural model (i.e., assuming the conditional expectation is linear). -

(Introduction to Probability at an Advanced Level) - All Lecture Notes
Fall 2018 Statistics 201A (Introduction to Probability at an advanced level) - All Lecture Notes Aditya Guntuboyina August 15, 2020 Contents 0.1 Sample spaces, Events, Probability.................................5 0.2 Conditional Probability and Independence.............................6 0.3 Random Variables..........................................7 1 Random Variables, Expectation and Variance8 1.1 Expectations of Random Variables.................................9 1.2 Variance................................................ 10 2 Independence of Random Variables 11 3 Common Distributions 11 3.1 Ber(p) Distribution......................................... 11 3.2 Bin(n; p) Distribution........................................ 11 3.3 Poisson Distribution......................................... 12 4 Covariance, Correlation and Regression 14 5 Correlation and Regression 16 6 Back to Common Distributions 16 6.1 Geometric Distribution........................................ 16 6.2 Negative Binomial Distribution................................... 17 7 Continuous Distributions 17 7.1 Normal or Gaussian Distribution.................................. 17 1 7.2 Uniform Distribution......................................... 18 7.3 The Exponential Density...................................... 18 7.4 The Gamma Density......................................... 18 8 Variable Transformations 19 9 Distribution Functions and the Quantile Transform 20 10 Joint Densities 22 11 Joint Densities under Transformations 23 11.1 Detour to Convolutions...................................... -

Large Scale Conditional Covariance Matrix Modeling, Estimation and Testing
Large Scale Conditional Covariance Matrix Modeling, Estimation and Testing Zhuanxin Ding1 Frank Russell Company, Tacoma, WA 98401, USA and Robert F. Engle University of California, San Diego, La Jolla, CA 92093, USA June 1996 This Revision May 2001 Abstract A new representation of the diagonal Vech model is given using the Hadamard product. Sufficient conditions on parameter matrices are provided to ensure the positive definiteness of covariance matrices from the new representation. Based on this, some new and simple models are discussed. A set of diagnostic tests for multivariate ARCH models is proposed. The tests are able to detect various model misspecifications by examing the orthogonality of the squared normalized residuals. A small Monte-Carlo study is carried out to check the small sample performance of the test. An empirical example is also given as guidance for model estimation and selection in the multivariate framework. For the specific data set considered, it is found that the simple one and two parameter models and the constant conditional correlation model perform fairly well. Keywords: conditional covariance, Multivariate ARCH, Hadamard product, M-test. 1 We would like to thank Clive Granger, Hong-Ye Gao, Doug Martin, Jeffrey Wang and Hal White for many helpful discussions. 2 1. Introduction It has now become common knowledge that the risk of the financial market, which is mainly represented by the variance of individual stock returns and the covariance between different assets or with the market, is highly forecastable. The research in this area however is far from complete and the application of different econometric models is just at its beginning. -

On the Autoregressive Conditional Heteroskedasticity Models
On the Autoregressive Conditional Heteroskedasticity Models By Erik Stenberg Department of Statistics Uppsala University Bachelor’s Thesis Supervisor: Yukai Yang 2016 Abstract This thesis aims to provide a thorough discussion on the early ARCH- type models, including a brief background with volatility clustering, kur- tosis, skewness of financial time series. In-sample fit of the widely used GARCH(1,1) is discussed using both standard model evaluation criteria and simulated values from estimated models. A rolling scheme out-of sam- ple forecast is evaluated using several different loss-functions and R2 from the Mincer-Zarnowitz regression, with daily realized volatility constructed using five minute intra-daily squared returns to proxy the true volatility. Keywords: ARCH, GARCH, Volatility clustering, fat tail, forecasting, intra-daily returns Contents 1 Introduction 3 2 Volatility 3 3 Autoregressive Conditional Heteroskedasticity 7 3.1 ARCH(1) . 8 3.2 Implied Kurtosis of ARCH(1) . 8 4 Generalized Autoregressive Conditional Heteroskedasticity 9 4.1 GARCH(1,1) . 9 5 Other Distributional Assumptions 11 5.1 Student-t . 11 5.2 Skewed Student-t . 12 5.3 Skewed Generalized Error Distribution . 12 6 Diagnostics 13 6.1 In-Sample Fit Evaluation . 13 6.2 Forecast Evaluation . 13 6.2.1 Value-at-Risk . 17 7 Illustrations 17 7.1 In-Sample Fit . 17 7.2 Forecast . 19 8 Summary, Conclusions and Further Research 21 9 Appendix 24 1 Introduction Ever since the first of many Autoregressive Conditional Heteroskedastic (ARCH) models was presented, see Engle (1982), fitting models to describe conditional heteroskedasticity has been a widely discussed topic. The main reason for this is the fact that up to that point, many of the conventional time series and models used in finance assumed a constant standard deviation in asset returns, see Black and Scholes (1973). -

Autoregressive Conditional Heteroscedasticity with Estimates of the Variance of United Kingdom Inflation Author(S): Robert F
Autoregressive Conditional Heteroscedasticity with Estimates of the Variance of United Kingdom Inflation Author(s): Robert F. Engle Source: Econometrica, Vol. 50, No. 4 (Jul., 1982), pp. 987-1007 Published by: The Econometric Society Stable URL: http://www.jstor.org/stable/1912773 Accessed: 06-11-2017 22:41 UTC REFERENCES Linked references are available on JSTOR for this article: http://www.jstor.org/stable/1912773?seq=1&cid=pdf-reference#references_tab_contents You may need to log in to JSTOR to access the linked references. JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at http://about.jstor.org/terms The Econometric Society is collaborating with JSTOR to digitize, preserve and extend access to Econometrica This content downloaded from 176.21.124.211 on Mon, 06 Nov 2017 22:41:38 UTC All use subject to http://about.jstor.org/terms Econometrica, Vol. 50, No. 4 (July, 1982) AUTOREGRESSIVE CONDITIONAL HETEROSCEDASTICITY WITH ESTIMATES OF THE VARIANCE OF UNITED KINGDOM INFLATION1 BY ROBERT F. ENGLE Traditional econometric models assume a constant one-period forecast variance. To generalize this implausible assumption, a new class of stochastic processes called autore- gressive conditional heteroscedastic (ARCH) processes are introduced in this paper. These are mean zero, serially uncorrelated processes with nonconstant variances conditional on the past, but constant unconditional variances. -

Lecture 21: Conditional Distributions and Covariance / Correlation
6.3, 6.4 Conditional Distributions Conditional Probability / Distributions Let X and Y be random variables then Lecture 21: Conditional Distributions and Covariance / Conditional probability: Correlation P(X = x; Y = y) P(X = xjY = y) = P(Y = y) Statistics 104 f (x; y) f (xjy) = Colin Rundel fY (y) Product rule: April 9, 2012 P(X = x; Y = y) = P(X = xjY = y)P(Y = y) f (x; y) = f (xjy)fY (y) Statistics 104 (Colin Rundel) Lecture 21 April 9, 2012 1 / 23 6.3, 6.4 Conditional Distributions 6.3, 6.4 Conditional Distributions Bayes' Theorem and the Law of Total Probability Example - Conditional Uniforms Let X and Y be random variables then Let X ∼ Unif(0; 1) and Y jX ∼ Unif(x; 1), find f (x; y), fY (y), and f (xjy). Bayes' Theorem: f (x; y) f (yjx)f (x) f (xjy) = = X fY (y) fY (y) f (x; y) f (xjy)f (y) f (yjx) = = Y fX (x) fX (x) Law of Total Probability: Z 1 Z 1 fX (x) = f (x; y) dy = f (xjy)fY (y) dy −∞ −∞ Z 1 Z 1 fY (y) = f (x; y) dx = f (yjx)fX (x) dx −∞ −∞ Statistics 104 (Colin Rundel) Lecture 21 April 9, 2012 2 / 23 Statistics 104 (Colin Rundel) Lecture 21 April 9, 2012 3 / 23 6.3, 6.4 Conditional Distributions 6.3, 6.4 Conditional Expectation Independence and Conditional Distributions Conditional Expectation If X and Y are independent random variables then If X and Y are independent random variables then we define the conditional expectation as follows f (xjy) = f (x) 4.7 Conditional Expectation X 257 X The value of E(Y x) will not be uniquely defined for those values of x such that E(X jY = y) = xf (xjy) dx f (they marginaljx) = p.f.fY or( p.d.f.y| ) of X satisfies f (x) 0. -

Heteroskedasticity in the Linear Model 2 Fall 2020 Unversit¨Atbasel
Short Guides to Microeconometrics Kurt Schmidheiny Heteroskedasticity in the Linear Model 2 Fall 2020 Unversit¨atBasel Note that under OLS2 (i.i.d. sample) the errors are unconditionally 2 homoscedastic, V [ui] = σ but allowed to be conditionally heteroscedas- tic, V [u jx ] = σ2. Assuming OLS2 and OLS3c provides that the errors Heteroskedasticity in the Linear Model i i i are also not conditionally autocorrelated, i.e. 8i 6= j : Cov[ui; ujjxi; xj] = E[uiujjxi; xj] = E[uijxi] · E[ujjxj] = 0 . Also note that the condition- ing on xi is less restrictive than it may seem: if the conditional variance V [uijxi] depends on other exogenous variables (or functions of them), we 1 Introduction can include these variables in xi and set the corresponding β parameters This handout extends the handout on \The Multiple Linear Regression to zero. 0 model" and refers to its definitions and assumptions in section 2. The variance-covariance of the vector of error terms u = (u1; u2; :::; uN ) This handouts relaxes the homoscedasticity assumption (OLS4a) and in the whole sample is therefore shows how the parameters of the linear model are correctly estimated and 0 2 tested when the error terms are heteroscedastic (OLS4b). V [ujX] = E[uu jX] = σ Ω 0 2 1 0 1 σ1 0 ··· 0 !1 0 ··· 0 2 The Econometric Model B 0 σ2 ··· 0 C B 0 ! ··· 0 C B 2 C 2 B 2 C = B . C = σ B . C B . .. C B . .. C Consider the multiple linear regression model for observation i = 1; :::; N @ . A @ . A 2 0 0 ··· σN 0 0 ··· !N 0 yi = xiβ + ui where x0 is a (K + 1)-dimensional row vector of K explanatory variables i 3 A Generic Case: Groupewise Heteroskedasticity and a constant, β is a (K + 1)-dimensional column vector of parameters and ui is a scalar called the error term. -

Autoregressive Conditional Heteroskedasticity Models
Chapter 5 Autoregressive Conditional Heteroskedasticity Models 5.1 Modeling Volatility In most econometric models the variance of the disturbance term is assumed to be constant (homoscedasticity). However, there is a number of economics and finance series that exhibit periods of unusual large volatility, followed by periods of relative tranquility. Then, in such cases the assumption of homoscedasticity is not appropri- ate. For example, consider the daily changes in the NYSE International 100 Index, April 5, 2004 - September 20, 2011, shown in Figure 5.1. One easy way to understand volatility is modeling it explicitly, for example in yt+1 = εt+1xt (5.1) where yt+1 is the variable of interest, εt+1 is a white-noise disturbance term with 2 variance σ , and xt is an independent variable that is observed at time t. If xt = xt 1 = xt 2 = = constant, then the yt is a familiar white-noise process − − ··· { } with a constant variance. However, if they are not constant, the variance of yt+1 conditional on the observable value of xt is 2 2 var (yt 1 xt ) = x σ (5.2) + | t If xt exhibit positive serial correlation, the conditional variance of the yt se- quence{ } will also exhibit positive serial correlation. We can write the model{ in loga-} rithm form and introduce the coefficients a0 and a1 to have log (yt ) = a0 + a1 log (xt 1) + et (5.3) − where et = log (εt ). 43 44 5 Autoregressive Conditional Heteroskedasticity Models 15 10 5 0 -5 Percent change Percent -10 -15 01jul2004 01jul2005 01jul2006 01jul2007 01jul2008 01jul2009 01jul2010 01jul2011 01jan2004 01jan2005 01jan2006 01jan2007 01jan2008 01jan2009 01jan2010 01jan2011 Date Fig. -

Arch — Autoregressive Conditional Heteroskedasticity (ARCH) Family of Estimators
Title stata.com arch — Autoregressive conditional heteroskedasticity (ARCH) family of estimators Description Quick start Menu Syntax Options Remarks and examples Stored results Methods and formulas References Also see Description arch fits regression models in which the volatility of a series varies through time. Usually, periods of high and low volatility are grouped together. ARCH models estimate future volatility as a function of prior volatility. To accomplish this, arch fits models of autoregressive conditional heteroskedasticity (ARCH) by using conditional maximum likelihood. In addition to ARCH terms, models may include multiplicative heteroskedasticity. Gaussian (normal), Student’s t, and generalized error distributions are supported. Concerning the regression equation itself, models may also contain ARCH-in-mean and ARMA terms. Quick start ARCH model of y with first- and second-order ARCH components and regressor x using tsset data arch y x, arch(1,2) Add a second-order GARCH component arch y x, arch(1,2) garch(2) Add an autoregressive component of order 2 and a moving-average component of order 3 arch y x, arch(1,2) garch(2) ar(2) ma(3) As above, but with the conditional variance included in the mean equation arch y x, arch(1,2) garch(2) ar(2) ma(3) archm EGARCH model of order 2 for y with an autoregressive component of order 1 arch y, earch(2) egarch(2) ar(1) 1 2 arch — Autoregressive conditional heteroskedasticity (ARCH) family of estimators Menu ARCH/GARCH Statistics > Time series > ARCH/GARCH > ARCH and GARCH models EARCH/EGARCH -

A Multivariate Variance Components Model for Analysis of Covariance In
Statistical Science 2009, Vol. 24, No. 2, 223–237 DOI: 10.1214/09-STS294 c Institute of Mathematical Statistics, 2009 A Multivariate Variance Components Model for Analysis of Covariance in Designed Experiments James G. Booth, Walter T. Federer, Martin T. Wells and Russell D. Wolfinger Abstract. Traditional methods for covariate adjustment of treatment means in designed experiments are inherently conditional on the ob- served covariate values. In order to develop a coherent general method- ology for analysis of covariance, we propose a multivariate variance components model for the joint distribution of the response and co- variates. It is shown that, if the design is orthogonal with respect to (random) blocking factors, then appropriate adjustments to treatment means can be made using the univariate variance components model obtained by conditioning on the observed covariate values. However, it is revealed that some widely used models are incorrectly specified, leading to biased estimates and incorrect standard errors. The approach clarifies some issues that have been the source of ongoing confusion in the statistics literature. Key words and phrases: Adjusted mean, blocking factor, conditional model, orthogonal design, randomized blocks design. 1. INTRODUCTION Cochran and Cox (1957), Snedecor and Cochran (1967), as well as more recent texts such as Milliken This article concerns the adjustment of treatment and Johnson (2002). We are specifically interested means in designed experiments to account for one or in settings in which the covariates are random vari- more covariates. Analysis of covariance has a long ables, not fixed by the design. Also, we generally history, dating back to Fisher (1934).