Fitting Linear Mixed-Effects Models Using Lme4
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lecture 22: Bivariate Normal Distribution Distribution
6.5 Conditional Distributions General Bivariate Normal Let Z1; Z2 ∼ N (0; 1), which we will use to build a general bivariate normal Lecture 22: Bivariate Normal Distribution distribution. 1 1 2 2 f (z1; z2) = exp − (z1 + z2 ) Statistics 104 2π 2 We want to transform these unit normal distributions to have the follow Colin Rundel arbitrary parameters: µX ; µY ; σX ; σY ; ρ April 11, 2012 X = σX Z1 + µX p 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 1 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - Marginals General Bivariate Normal - Cov/Corr First, lets examine the marginal distributions of X and Y , Second, we can find Cov(X ; Y ) and ρ(X ; Y ) Cov(X ; Y ) = E [(X − E(X ))(Y − E(Y ))] X = σX Z1 + µX h p i = E (σ Z + µ − µ )(σ [ρZ + 1 − ρ2Z ] + µ − µ ) = σX N (0; 1) + µX X 1 X X Y 1 2 Y Y 2 h p 2 i = N (µX ; σX ) = E (σX Z1)(σY [ρZ1 + 1 − ρ Z2]) h 2 p 2 i = σX σY E ρZ1 + 1 − ρ Z1Z2 p 2 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY = σX σY ρE[Z1 ] p 2 = σX σY ρ = σY [ρN (0; 1) + 1 − ρ N (0; 1)] + µY = σ [N (0; ρ2) + N (0; 1 − ρ2)] + µ Y Y Cov(X ; Y ) ρ(X ; Y ) = = ρ = σY N (0; 1) + µY σX σY 2 = N (µY ; σY ) Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 2 / 22 Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 3 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - RNG Multivariate Change of Variables Consequently, if we want to generate a Bivariate Normal random variable Let X1;:::; Xn have a continuous joint distribution with pdf f defined of S. -

Linear Mixed-Effects Modeling in SPSS: an Introduction to the MIXED Procedure
Technical report Linear Mixed-Effects Modeling in SPSS: An Introduction to the MIXED Procedure Table of contents Introduction. 1 Data preparation for MIXED . 1 Fitting fixed-effects models . 4 Fitting simple mixed-effects models . 7 Fitting mixed-effects models . 13 Multilevel analysis . 16 Custom hypothesis tests . 18 Covariance structure selection. 19 Random coefficient models . 20 Estimated marginal means. 25 References . 28 About SPSS Inc. 28 SPSS is a registered trademark and the other SPSS products named are trademarks of SPSS Inc. All other names are trademarks of their respective owners. © 2005 SPSS Inc. All rights reserved. LMEMWP-0305 Introduction The linear mixed-effects models (MIXED) procedure in SPSS enables you to fit linear mixed-effects models to data sampled from normal distributions. Recent texts, such as those by McCulloch and Searle (2000) and Verbeke and Molenberghs (2000), comprehensively review mixed-effects models. The MIXED procedure fits models more general than those of the general linear model (GLM) procedure and it encompasses all models in the variance components (VARCOMP) procedure. This report illustrates the types of models that MIXED handles. We begin with an explanation of simple models that can be fitted using GLM and VARCOMP, to show how they are translated into MIXED. We then proceed to fit models that are unique to MIXED. The major capabilities that differentiate MIXED from GLM are that MIXED handles correlated data and unequal variances. Correlated data are very common in such situations as repeated measurements of survey respondents or experimental subjects. MIXED extends repeated measures models in GLM to allow an unequal number of repetitions. -

Application of Random-Effects Probit Regression Models
Journal of Consulting and Clinical Psychology Copyright 1994 by the American Psychological Association, Inc. 1994, Vol. 62, No. 2, 285-296 0022-006X/94/S3.00 Application of Random-Effects Probit Regression Models Robert D. Gibbons and Donald Hedeker A random-effects probit model is developed for the case in which the outcome of interest is a series of correlated binary responses. These responses can be obtained as the product of a longitudinal response process where an individual is repeatedly classified on a binary outcome variable (e.g., sick or well on occasion t), or in "multilevel" or "clustered" problems in which individuals within groups (e.g., firms, classes, families, or clinics) are considered to share characteristics that produce similar responses. Both examples produce potentially correlated binary responses and modeling these per- son- or cluster-specific effects is required. The general model permits analysis at both the level of the individual and cluster and at the level at which experimental manipulations are applied (e.g., treat- ment group). The model provides maximum likelihood estimates for time-varying and time-invari- ant covariates in the longitudinal case and covariates which vary at the level of the individual and at the cluster level for multilevel problems. A similar number of individuals within clusters or number of measurement occasions within individuals is not required. Empirical Bayesian estimates of per- son-specific trends or cluster-specific effects are provided. Models are illustrated with data from -

Multilevel and Mixed-Effects Modeling 391
386 Statistics with Stata , . d itl 770/ ""01' ncdctemp However the residuals pass tests for white noise 111uahtemp, compar e WI 1 /0 l' r : , , '12 19) (p = .65), and a plot of observed and predicted values shows a good visual fit (Figure . , Multilevel and Mixed-Effects predict uahhat2 solar, ENSO & C02" label variable uahhat2 "predicted from volcanoes, Modeling predict uahres2, resid label variable uahres2 "UAH residuals from ARMAX model" wntestq uahres2, lags(25) portmanteau test for white noise portmanteau (Q)statistic 21. 7197 Mixed-effects modeling is basically regression analysis allowing two kinds of effects: fixed =rob > chi2(25) 0.6519 effects, meaning intercepts and slopes meant to describe the population as a whole, just as in Figure 12.19 ordinary regression; and also random effects, meaning intercepts and slopes that can vary across subgroups of the sample. All of the regression-type methods shown so far in this book involve fixed effects only. Mixed-effects modeling opens a new range of possibilities for multilevel o models, growth curve analysis, and panel data or cross-sectional time series, "r~ 00 01 Albright and Marinova (2010) provide a practical comparison of mixed-modeling procedures uj"'! > found in Stata, SAS, SPSS and R with the hierarchical linear modeling (HLM) software 0>- m developed by Raudenbush and Bryck (2002; also Raudenbush et al. 2005). More detailed ~o c explanation of mixed modeling and its correspondences with HLM can be found in Rabe• ro (I! Hesketh and Skrondal (2012). Briefly, HLM approaches multilevel modeling in several steps, ::IN co I' specifying separate equations (for example) for levelland level 2 effects. -

10.1 Topic 10. ANOVA Models for Random and Mixed Effects References
10.1 Topic 10. ANOVA models for random and mixed effects References: ST&DT: Topic 7.5 p.152-153, Topic 9.9 p. 225-227, Topic 15.5 379-384. There is a good discussion in SAS System for Linear Models, 3rd ed. pages 191-198. Note: do not use the rules for expected MS on ST&D page 381. We provide in the notes below updated rules from Chapter 8 from Montgomery D.C. (1991) “Design and analysis of experiments”. 10. 1. Introduction The experiments discussed in previous chapters have dealt primarily with situations in which the experimenter is concerned with making comparisons among specific factor levels. There are other types of experiments, however, in which one wish to determine the sources of variation in a system rather than make particular comparisons. One may also wish to extend the conclusions beyond the set of specific factor levels included in the experiment. The purpose of this chapter is to introduce analysis of variance models that are appropriate to these different kinds of experimental objectives. 10. 2. Fixed and Random Models in One-way Classification Experiments 10. 2. 1. Fixed-effects model A typical fixed-effect analysis of an experiment involves comparing treatment means to one another in an attempt to detect differences. For example, four different forms of nitrogen fertilizer are compared in a CRD with five replications. The analysis of variance takes the following familiar form: Source df Total 19 Treatment (i.e. among groups) 3 Error (i.e. within groups) 16 And the linear model for this experiment is: Yij = µ + i + ij. -

Mixed Model Methodology, Part I: Linear Mixed Models
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/271372856 Mixed Model Methodology, Part I: Linear Mixed Models Technical Report · January 2015 DOI: 10.13140/2.1.3072.0320 CITATIONS READS 0 444 1 author: jean-louis Foulley Université de Montpellier 313 PUBLICATIONS 4,486 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Football Predictions View project All content following this page was uploaded by jean-louis Foulley on 27 January 2015. The user has requested enhancement of the downloaded file. Jean-Louis Foulley Mixed Model Methodology 1 Jean-Louis Foulley Institut de Mathématiques et de Modélisation de Montpellier (I3M) Université de Montpellier 2 Sciences et Techniques Place Eugène Bataillon 34095 Montpellier Cedex 05 2 Part I Linear Mixed Model Models Citation Foulley, J.L. (2015). Mixed Model Methodology. Part I: Linear Mixed Models. Technical Report, e-print: DOI: 10.13140/2.1.3072.0320 3 1 Basics on linear models 1.1 Introduction Linear models form one of the most widely used tools of statistics both from a theoretical and practical points of view. They encompass regression and analysis of variance and covariance, and presenting them within a unique theoretical framework helps to clarify the basic concepts and assumptions underlying all these techniques and the ones that will be used later on for mixed models. There is a large amount of highly documented literature especially textbooks in this area from both theoretical (Searle, 1971, 1987; Rao, 1973; Rao et al., 2007) and practical points of view (Littell et al., 2002). -
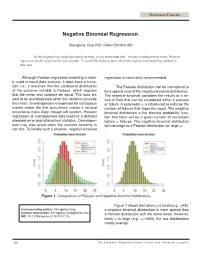
Negative Binomial Regression
STATISTICS COLUMN Negative Binomial Regression Shengping Yang PhD ,Gilbert Berdine MD In the hospital stay study discussed recently, it was mentioned that “in case overdispersion exists, Poisson regression model might not be appropriate.” I would like to know more about the appropriate modeling method in that case. Although Poisson regression modeling is wide- regression is commonly recommended. ly used in count data analysis, it does have a limita- tion, i.e., it assumes that the conditional distribution The Poisson distribution can be considered to of the outcome variable is Poisson, which requires be a special case of the negative binomial distribution. that the mean and variance be equal. The data are The negative binomial considers the results of a se- said to be overdispersed when the variance exceeds ries of trials that can be considered either a success the mean. Overdispersion is expected for contagious or failure. A parameter ψ is introduced to indicate the events where the first occurrence makes a second number of failures that stops the count. The negative occurrence more likely, though still random. Poisson binomial distribution is the discrete probability func- regression of overdispersed data leads to a deflated tion that there will be a given number of successes standard error and inflated test statistics. Overdisper- before ψ failures. The negative binomial distribution sion may also result when the success outcome is will converge to a Poisson distribution for large ψ. not rare. To handle such a situation, negative binomial 0 5 10 15 20 25 0 5 10 15 20 25 Figure 1. Comparison of Poisson and negative binomial distributions. -

Section 1: Regression Review
Section 1: Regression review Yotam Shem-Tov Fall 2015 Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 1 / 58 Contact information Yotam Shem-Tov, PhD student in economics. E-mail: [email protected]. Office: Evans hall room 650 Office hours: To be announced - any preferences or constraints? Feel free to e-mail me. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 2 / 58 R resources - Prior knowledge in R is assumed In the link here is an excellent introduction to statistics in R . There is a free online course in coursera on programming in R. Here is a link to the course. An excellent book for implementing econometric models and method in R is Applied Econometrics with R. This is my favourite for starting to learn R for someone who has some background in statistic methods in the social science. The book Regression Models for Data Science in R has a nice online version. It covers the implementation of regression models using R . A great link to R resources and other cool programming and econometric tools is here. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 3 / 58 Outline There are two general approaches to regression 1 Regression as a model: a data generating process (DGP) 2 Regression as an algorithm (i.e., an estimator without assuming an underlying structural model). Overview of this slides: 1 The conditional expectation function - a motivation for using OLS. 2 OLS as the minimum mean squared error linear approximation (projections). 3 Regression as a structural model (i.e., assuming the conditional expectation is linear). -

Chapter 17 Random Effects and Mixed Effects Models
Chapter 17 Random Effects and Mixed Effects Models Example. Solutions of alcohol are used for calibrating Breathalyzers. The following data show the alcohol concentrations of samples of alcohol solutions taken from six bottles of alcohol solution randomly selected from a large batch. The objective is to determine if all bottles in the batch have the same alcohol concentrations. Bottle Concentration 1 1.4357 1.4348 1.4336 1.4309 2 1.4244 1.4232 1.4213 1.4256 3 1.4153 1.4137 1.4176 1.4164 4 1.4331 1.4325 1.4312 1.4297 5 1.4252 1.4261 1.4293 1.4272 6 1.4179 1.4217 1.4191 1.4204 We are not interested in any difference between the six bottles used in the experiment. We therefore treat the six bottles as a random sample from the population and use a random effects model. If we were interested in the six bottles, we would use a fixed effects model. 17.3. One Random Factor 17.3.1. The Random‐Effects One‐way Model For a completely randomized design, with v treatments of a factor T, the one‐way random effects model is , :0, , :0, , all random variables are independent, i=1, ..., v, t=1, ..., . Note now , and are called variance components. Our interest is to investigate the variation of treatment effects. In a fixed effect model, we can test whether treatment effects are equal. In a random effects model, instead of testing : ..., against : treatment effects not all equal, we use : 0,: 0. If =0, then all random effects are 0. -

(Introduction to Probability at an Advanced Level) - All Lecture Notes
Fall 2018 Statistics 201A (Introduction to Probability at an advanced level) - All Lecture Notes Aditya Guntuboyina August 15, 2020 Contents 0.1 Sample spaces, Events, Probability.................................5 0.2 Conditional Probability and Independence.............................6 0.3 Random Variables..........................................7 1 Random Variables, Expectation and Variance8 1.1 Expectations of Random Variables.................................9 1.2 Variance................................................ 10 2 Independence of Random Variables 11 3 Common Distributions 11 3.1 Ber(p) Distribution......................................... 11 3.2 Bin(n; p) Distribution........................................ 11 3.3 Poisson Distribution......................................... 12 4 Covariance, Correlation and Regression 14 5 Correlation and Regression 16 6 Back to Common Distributions 16 6.1 Geometric Distribution........................................ 16 6.2 Negative Binomial Distribution................................... 17 7 Continuous Distributions 17 7.1 Normal or Gaussian Distribution.................................. 17 1 7.2 Uniform Distribution......................................... 18 7.3 The Exponential Density...................................... 18 7.4 The Gamma Density......................................... 18 8 Variable Transformations 19 9 Distribution Functions and the Quantile Transform 20 10 Joint Densities 22 11 Joint Densities under Transformations 23 11.1 Detour to Convolutions...................................... -

R2 Statistics for Mixed Models
Kansas State University Libraries New Prairie Press Conference on Applied Statistics in Agriculture 2005 - 17th Annual Conference Proceedings R2 STATISTICS FOR MIXED MODELS Matthew Kramer Follow this and additional works at: https://newprairiepress.org/agstatconference Part of the Agriculture Commons, and the Applied Statistics Commons This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 4.0 License. Recommended Citation Kramer, Matthew (2005). "R2 STATISTICS FOR MIXED MODELS," Conference on Applied Statistics in Agriculture. https://doi.org/10.4148/2475-7772.1142 This is brought to you for free and open access by the Conferences at New Prairie Press. It has been accepted for inclusion in Conference on Applied Statistics in Agriculture by an authorized administrator of New Prairie Press. For more information, please contact [email protected]. Conference on Applied Statistics in Agriculture Kansas State University 148 Kansas State University R2 STATISTICS FOR MIXED MODELS Matthew Kramer Biometrical Consulting Service, ARS (Beltsville, MD), USDA Abstract The R2 statistic, when used in a regression or ANOVA context, is appealing because it summarizes how well the model explains the data in an easy-to- understand way. R2 statistics are also useful to gauge the effect of changing a model. Generalizing R2 to mixed models is not obvious when there are correlated errors, as might occur if data are georeferenced or result from a designed experiment with blocking. Such an R2 statistic might refer only to the explanation associated with the independent variables, or might capture the explanatory power of the whole model. In the latter case, one might develop an R2 statistic from Wald or likelihood ratio statistics, but these can yield different numeric results. -

Linear Mixed Models and Penalized Least Squares
Linear mixed models and penalized least squares Douglas M. Bates Department of Statistics, University of Wisconsin–Madison Saikat DebRoy Department of Biostatistics, Harvard School of Public Health Abstract Linear mixed-effects models are an important class of statistical models that are not only used directly in many fields of applications but also used as iterative steps in fitting other types of mixed-effects models, such as generalized linear mixed models. The parameters in these models are typically estimated by maximum likelihood (ML) or restricted maximum likelihood (REML). In general there is no closed form solution for these estimates and they must be determined by iterative algorithms such as EM iterations or general nonlinear optimization. Many of the intermediate calculations for such iterations have been expressed as generalized least squares problems. We show that an alternative representation as a penalized least squares problem has many advantageous computational properties including the ability to evaluate explicitly a profiled log-likelihood or log-restricted likelihood, the gradient and Hessian of this profiled objective, and an ECME update to refine this objective. Key words: REML, gradient, Hessian, EM algorithm, ECME algorithm, maximum likelihood, profile likelihood, multilevel models 1 Introduction We will establish some results for the penalized least squares representation of a general form of a linear mixed-effects model, then show how these results specialize for particular models. The general form we consider is y = Xβ + Zb + ∼ N (0, σ2I), b ∼ N (0, σ2Ω−1), ⊥ b (1) where y is the n-dimensional response vector, X is an n × p model matrix for the p-dimensional fixed-effects vector β, Z is the n × q model matrix for Preprint submitted to Journal of Multivariate Analysis 25 September 2003 the q-dimensional random-effects vector b that has a Gaussian distribution with mean 0 and relative precision matrix Ω (i.e., Ω is the precision of b relative to the precision of ), and is the random noise assumed to have a spherical Gaussian distribution.