Lecture 7 Count Data Models
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lecture 22: Bivariate Normal Distribution Distribution
6.5 Conditional Distributions General Bivariate Normal Let Z1; Z2 ∼ N (0; 1), which we will use to build a general bivariate normal Lecture 22: Bivariate Normal Distribution distribution. 1 1 2 2 f (z1; z2) = exp − (z1 + z2 ) Statistics 104 2π 2 We want to transform these unit normal distributions to have the follow Colin Rundel arbitrary parameters: µX ; µY ; σX ; σY ; ρ April 11, 2012 X = σX Z1 + µX p 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 1 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - Marginals General Bivariate Normal - Cov/Corr First, lets examine the marginal distributions of X and Y , Second, we can find Cov(X ; Y ) and ρ(X ; Y ) Cov(X ; Y ) = E [(X − E(X ))(Y − E(Y ))] X = σX Z1 + µX h p i = E (σ Z + µ − µ )(σ [ρZ + 1 − ρ2Z ] + µ − µ ) = σX N (0; 1) + µX X 1 X X Y 1 2 Y Y 2 h p 2 i = N (µX ; σX ) = E (σX Z1)(σY [ρZ1 + 1 − ρ Z2]) h 2 p 2 i = σX σY E ρZ1 + 1 − ρ Z1Z2 p 2 2 Y = σY [ρZ1 + 1 − ρ Z2] + µY = σX σY ρE[Z1 ] p 2 = σX σY ρ = σY [ρN (0; 1) + 1 − ρ N (0; 1)] + µY = σ [N (0; ρ2) + N (0; 1 − ρ2)] + µ Y Y Cov(X ; Y ) ρ(X ; Y ) = = ρ = σY N (0; 1) + µY σX σY 2 = N (µY ; σY ) Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 2 / 22 Statistics 104 (Colin Rundel) Lecture 22 April 11, 2012 3 / 22 6.5 Conditional Distributions 6.5 Conditional Distributions General Bivariate Normal - RNG Multivariate Change of Variables Consequently, if we want to generate a Bivariate Normal random variable Let X1;:::; Xn have a continuous joint distribution with pdf f defined of S. -

An Introduction to Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A
NESUG 2010 Statistics and Analysis An Animated Guide: An Introduction To Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A. ABSTRACT: This paper will be a brief introduction to Poisson regression (theory, steps to be followed, complications and interpretation) via a worked example. It is hoped that this will increase motivation towards learning this useful statistical technique. INTRODUCTION: Poisson regression is available in SAS through the GENMOD procedure (generalized modeling). It is appropriate when: 1) the process that generates the conditional Y distributions would, theoretically, be expected to be a Poisson random process and 2) when there is no evidence of overdispersion and 3) when the mean of the marginal distribution is less than ten (preferably less than five and ideally close to one). THE POISSON DISTRIBUTION: The Poison distribution is a discrete Percent of observations where the random variable X is expected distribution and is appropriate for to have the value x, given that the Poisson distribution has a mean modeling counts of observations. of λ= P(X=x, λ ) = (e - λ * λ X) / X! Counts are observed cases, like the 0.4 count of measles cases in cities. You λ can simply model counts if all data 0.35 were collected in the same measuring 0.3 unit (e.g. the same number of days or 0.3 0.5 0.8 same number of square feet). 0.25 λ 1 0.2 3 You can use the Poisson Distribution = 5 for modeling rates (rates are counts 0.15 20 per unit) if the units of collection were 8 different. -

Generalized Linear Models (Glms)
San Jos´eState University Math 261A: Regression Theory & Methods Generalized Linear Models (GLMs) Dr. Guangliang Chen This lecture is based on the following textbook sections: • Chapter 13: 13.1 – 13.3 Outline of this presentation: • What is a GLM? • Logistic regression • Poisson regression Generalized Linear Models (GLMs) What is a GLM? In ordinary linear regression, we assume that the response is a linear function of the regressors plus Gaussian noise: 0 2 y = β0 + β1x1 + ··· + βkxk + ∼ N(x β, σ ) | {z } |{z} linear form x0β N(0,σ2) noise The model can be reformulate in terms of • distribution of the response: y | x ∼ N(µ, σ2), and • dependence of the mean on the predictors: µ = E(y | x) = x0β Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University3/24 Generalized Linear Models (GLMs) beta=(1,2) 5 4 3 β0 + β1x b y 2 y 1 0 −1 0.0 0.2 0.4 0.6 0.8 1.0 x x Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University4/24 Generalized Linear Models (GLMs) Generalized linear models (GLM) extend linear regression by allowing the response variable to have • a general distribution (with mean µ = E(y | x)) and • a mean that depends on the predictors through a link function g: That is, g(µ) = β0x or equivalently, µ = g−1(β0x) Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University5/24 Generalized Linear Models (GLMs) In GLM, the response is typically assumed to have a distribution in the exponential family, which is a large class of probability distributions that have pdfs of the form f(x | θ) = a(x)b(θ) exp(c(θ) · T (x)), including • Normal - ordinary linear regression • Bernoulli - Logistic regression, modeling binary data • Binomial - Multinomial logistic regression, modeling general cate- gorical data • Poisson - Poisson regression, modeling count data • Exponential, Gamma - survival analysis Dr. -

Generalized Linear Models with Poisson Family: Applications in Ecology
UNIVERSITY OF ABOMEY- CALAVI *********** FACULTY OF AGRONOMIC SCIENCES *************** **************** Master Program in Statistics, Major Biostatistics 1st batch Generalized linear models with Poisson family: applications in ecology A thesis submitted to the Faculty of Agronomic Sciences in partial fulfillment of the requirements for the degree of the Master of Sciences in Biostatistics Presented by: LOKONON Enagnon Bruno Supervisor: Pr Romain L. GLELE KAKAÏ, Professor of Biostatistics and Forest estimation Academic year: 2014-2015 UNIVERSITE D’ABOMEY- CALAVI *********** FACULTE DES SCIENCES AGRONOMIQUES *************** ************** Programme de Master en Biostatistiques 1ère Promotion Modèles linéaires généralisés de la famille de Poisson : applications en écologie Mémoire soumis à la Faculté des Sciences Agronomiques pour obtenir le Diplôme de Master recherche en Biostatistiques Présenté par: LOKONON Enagnon Bruno Superviseur: Pr Romain L. GLELE KAKAÏ, Professeur titulaire de Biostatistiques et estimation forestière Année académique: 2014-2015 Certification I certify that this work has been achieved by LOKONON E. Bruno under my entire supervision at the University of Abomey-Calavi (Benin) in order to obtain his Master of Science degree in Biostatistics. Pr Romain L. GLELE KAKAÏ Professor of Biostatistics and Forest estimation i Acknowledgements This research was supported by WAAPP/PPAAO-BENIN (West African Agricultural Productivity Program/ Programme de Productivité Agricole en Afrique de l‟Ouest). This dissertation could only have been possible through the generous contributions of many people. First and foremost, I am grateful to my supervisor Pr Romain L. GLELE KAKAÏ, Professor of Biostatistics and Forest estimation who tirelessly played key role in orientation, scientific writing and mentoring during this research. In particular, I thank him for his prompt availability whenever needed. -

Parametric Models
An Introduction to Event History Analysis Oxford Spring School June 18-20, 2007 Day Two: Regression Models for Survival Data Parametric Models We’ll spend the morning introducing regression-like models for survival data, starting with fully parametric (distribution-based) models. These tend to be very widely used in social sciences, although they receive almost no use outside of that (e.g., in biostatistics). A General Framework (That Is, Some Math) Parametric models are continuous-time models, in that they assume a continuous parametric distribution for the probability of failure over time. A general parametric duration model takes as its starting point the hazard: f(t) h(t) = (1) S(t) As we discussed yesterday, the density is the probability of the event (at T ) occurring within some differentiable time-span: Pr(t ≤ T < t + ∆t) f(t) = lim . (2) ∆t→0 ∆t The survival function is equal to one minus the CDF of the density: S(t) = Pr(T ≥ t) Z t = 1 − f(t) dt 0 = 1 − F (t). (3) This means that another way of thinking of the hazard in continuous time is as a conditional limit: Pr(t ≤ T < t + ∆t|T ≥ t) h(t) = lim (4) ∆t→0 ∆t i.e., the conditional probability of the event as ∆t gets arbitrarily small. 1 A General Parametric Likelihood For a set of observations indexed by i, we can distinguish between those which are censored and those which aren’t... • Uncensored observations (Ci = 1) tell us both about the hazard of the event, and the survival of individuals prior to that event. -

Overdispersion, and How to Deal with It in R and JAGS (Requires R-Packages AER, Coda, Lme4, R2jags, Dharma/Devtools) Carsten F
Overdispersion, and how to deal with it in R and JAGS (requires R-packages AER, coda, lme4, R2jags, DHARMa/devtools) Carsten F. Dormann 07 December, 2016 Contents 1 Introduction: what is overdispersion? 1 2 Recognising (and testing for) overdispersion 1 3 “Fixing” overdispersion 5 3.1 Quasi-families . .5 3.2 Different distribution (here: negative binomial) . .6 3.3 Observation-level random effects (OLRE) . .8 4 Overdispersion in JAGS 12 1 Introduction: what is overdispersion? Overdispersion describes the observation that variation is higher than would be expected. Some distributions do not have a parameter to fit variability of the observation. For example, the normal distribution does that through the parameter σ (i.e. the standard deviation of the model), which is constant in a typical regression. In contrast, the Poisson distribution has no such parameter, and in fact the variance increases with the mean (i.e. the variance and the mean have the same value). In this latter case, for an expected value of E(y) = 5, we also expect that the variance of observed data points is 5. But what if it is not? What if the observed variance is much higher, i.e. if the data are overdispersed? (Note that it could also be lower, underdispersed. This is less often the case, and not all approaches below allow for modelling underdispersion, but some do.) Overdispersion arises in different ways, most commonly through “clumping”. Imagine the number of seedlings in a forest plot. Depending on the distance to the source tree, there may be many (hundreds) or none. The same goes for shooting stars: either the sky is empty, or littered with shooting stars. -

Poisson Versus Negative Binomial Regression
Handling Count Data The Negative Binomial Distribution Other Applications and Analysis in R References Poisson versus Negative Binomial Regression Randall Reese Utah State University [email protected] February 29, 2016 Randall Reese Poisson and Neg. Binom Handling Count Data The Negative Binomial Distribution Other Applications and Analysis in R References Overview 1 Handling Count Data ADEM Overdispersion 2 The Negative Binomial Distribution Foundations of Negative Binomial Distribution Basic Properties of the Negative Binomial Distribution Fitting the Negative Binomial Model 3 Other Applications and Analysis in R 4 References Randall Reese Poisson and Neg. Binom Handling Count Data The Negative Binomial Distribution ADEM Other Applications and Analysis in R Overdispersion References Count Data Randall Reese Poisson and Neg. Binom Handling Count Data The Negative Binomial Distribution ADEM Other Applications and Analysis in R Overdispersion References Count Data Data whose values come from Z≥0, the non-negative integers. Classic example is deaths in the Prussian army per year by horse kick (Bortkiewicz) Example 2 of Notes 5. (Number of successful \attempts"). Randall Reese Poisson and Neg. Binom Handling Count Data The Negative Binomial Distribution ADEM Other Applications and Analysis in R Overdispersion References Poisson Distribution Support is the non-negative integers. (Count data). Described by a single parameter λ > 0. When Y ∼ Poisson(λ), then E(Y ) = Var(Y ) = λ Randall Reese Poisson and Neg. Binom Handling Count Data The Negative Binomial Distribution ADEM Other Applications and Analysis in R Overdispersion References Acute Disseminated Encephalomyelitis Acute Disseminated Encephalomyelitis (ADEM) is a neurological, immune disorder in which widespread inflammation of the brain and spinal cord damages tissue known as white matter. -
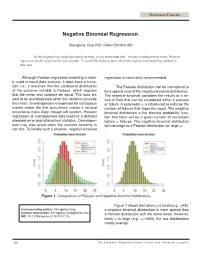
Negative Binomial Regression
STATISTICS COLUMN Negative Binomial Regression Shengping Yang PhD ,Gilbert Berdine MD In the hospital stay study discussed recently, it was mentioned that “in case overdispersion exists, Poisson regression model might not be appropriate.” I would like to know more about the appropriate modeling method in that case. Although Poisson regression modeling is wide- regression is commonly recommended. ly used in count data analysis, it does have a limita- tion, i.e., it assumes that the conditional distribution The Poisson distribution can be considered to of the outcome variable is Poisson, which requires be a special case of the negative binomial distribution. that the mean and variance be equal. The data are The negative binomial considers the results of a se- said to be overdispersed when the variance exceeds ries of trials that can be considered either a success the mean. Overdispersion is expected for contagious or failure. A parameter ψ is introduced to indicate the events where the first occurrence makes a second number of failures that stops the count. The negative occurrence more likely, though still random. Poisson binomial distribution is the discrete probability func- regression of overdispersed data leads to a deflated tion that there will be a given number of successes standard error and inflated test statistics. Overdisper- before ψ failures. The negative binomial distribution sion may also result when the success outcome is will converge to a Poisson distribution for large ψ. not rare. To handle such a situation, negative binomial 0 5 10 15 20 25 0 5 10 15 20 25 Figure 1. Comparison of Poisson and negative binomial distributions. -

Statistical Approaches for Highly Skewed Data 1
Running head: STATISTICAL APPROACHES FOR HIGHLY SKEWED DATA 1 (in press) Journal of Clinical Child and Adolescent Psychology Statistical Approaches for Highly Skewed Data: Evaluating Relations between Child Maltreatment and Young Adults’ Non-Suicidal Self-injury Ana Gonzalez-Blanks, PhD Jessie M. Bridgewater, MA *Tuppett M. Yates, PhD Department of Psychology University of California, Riverside Acknowledgments: We gratefully acknowledge Dr. Chandra Reynolds who provided generous instruction and consultation throughout the execution of the analyses reported herein. *Please address correspondence to: Tuppett M. Yates, Department of Psychology, University of California, Riverside CA, 92521. Email: [email protected]. STATISTICAL APPROACHES FOR HIGHLY SKEWED DATA 2 Abstract Objective: Clinical phenomena often feature skewed distributions with an overabundance of zeros. Unfortunately, empirical methods for dealing with this violation of distributional assumptions underlying regression are typically discussed in statistical journals with limited translation to applied researchers. Therefore, this investigation compared statistical approaches for addressing highly skewed data as applied to the evaluation of relations between child maltreatment and non-suicidal self-injury (NSSI). Method: College students (N = 2,651; 64.2% female; 85.2% non-White) completed the Child Abuse and Trauma Scale and the Functional Assessment of Self-Mutilation. Statistical models were applied to cross-sectional data to provide illustrative comparisons across predictions to a) raw, highly skewed NSSI outcomes, b) natural log, square-root, and inverse NSSI transformations to reduce skew, c) zero-inflated Poisson (ZIP) and negative-binomial zero- inflated (NBZI) regression models to account for both disproportionate zeros and skewness in the NSSI data, and d) the skew-t distribution to model NSSI skewness. -

Section 1: Regression Review
Section 1: Regression review Yotam Shem-Tov Fall 2015 Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 1 / 58 Contact information Yotam Shem-Tov, PhD student in economics. E-mail: [email protected]. Office: Evans hall room 650 Office hours: To be announced - any preferences or constraints? Feel free to e-mail me. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 2 / 58 R resources - Prior knowledge in R is assumed In the link here is an excellent introduction to statistics in R . There is a free online course in coursera on programming in R. Here is a link to the course. An excellent book for implementing econometric models and method in R is Applied Econometrics with R. This is my favourite for starting to learn R for someone who has some background in statistic methods in the social science. The book Regression Models for Data Science in R has a nice online version. It covers the implementation of regression models using R . A great link to R resources and other cool programming and econometric tools is here. Yotam Shem-Tov STAT 239A/ PS 236A September 3, 2015 3 / 58 Outline There are two general approaches to regression 1 Regression as a model: a data generating process (DGP) 2 Regression as an algorithm (i.e., an estimator without assuming an underlying structural model). Overview of this slides: 1 The conditional expectation function - a motivation for using OLS. 2 OLS as the minimum mean squared error linear approximation (projections). 3 Regression as a structural model (i.e., assuming the conditional expectation is linear). -

Negative Binomial Regression Models and Estimation Methods
Appendix D: Negative Binomial Regression Models and Estimation Methods By Dominique Lord Texas A&M University Byung-Jung Park Korea Transport Institute This appendix presents the characteristics of Negative Binomial regression models and discusses their estimating methods. Probability Density and Likelihood Functions The properties of the negative binomial models with and without spatial intersection are described in the next two sections. Poisson-Gamma Model The Poisson-Gamma model has properties that are very similar to the Poisson model discussed in Appendix C, in which the dependent variable yi is modeled as a Poisson variable with a mean i where the model error is assumed to follow a Gamma distribution. As it names implies, the Poisson-Gamma is a mixture of two distributions and was first derived by Greenwood and Yule (1920). This mixture distribution was developed to account for over-dispersion that is commonly observed in discrete or count data (Lord et al., 2005). It became very popular because the conjugate distribution (same family of functions) has a closed form and leads to the negative binomial distribution. As discussed by Cook (2009), “the name of this distribution comes from applying the binomial theorem with a negative exponent.” There are two major parameterizations that have been proposed and they are known as the NB1 and NB2, the latter one being the most commonly known and utilized. NB2 is therefore described first. Other parameterizations exist, but are not discussed here (see Maher and Summersgill, 1996; Hilbe, 2007). NB2 Model Suppose that we have a series of random counts that follows the Poisson distribution: i e i gyii; (D-1) yi ! where yi is the observed number of counts for i 1, 2, n ; and i is the mean of the Poisson distribution. -

(Introduction to Probability at an Advanced Level) - All Lecture Notes
Fall 2018 Statistics 201A (Introduction to Probability at an advanced level) - All Lecture Notes Aditya Guntuboyina August 15, 2020 Contents 0.1 Sample spaces, Events, Probability.................................5 0.2 Conditional Probability and Independence.............................6 0.3 Random Variables..........................................7 1 Random Variables, Expectation and Variance8 1.1 Expectations of Random Variables.................................9 1.2 Variance................................................ 10 2 Independence of Random Variables 11 3 Common Distributions 11 3.1 Ber(p) Distribution......................................... 11 3.2 Bin(n; p) Distribution........................................ 11 3.3 Poisson Distribution......................................... 12 4 Covariance, Correlation and Regression 14 5 Correlation and Regression 16 6 Back to Common Distributions 16 6.1 Geometric Distribution........................................ 16 6.2 Negative Binomial Distribution................................... 17 7 Continuous Distributions 17 7.1 Normal or Gaussian Distribution.................................. 17 1 7.2 Uniform Distribution......................................... 18 7.3 The Exponential Density...................................... 18 7.4 The Gamma Density......................................... 18 8 Variable Transformations 19 9 Distribution Functions and the Quantile Transform 20 10 Joint Densities 22 11 Joint Densities under Transformations 23 11.1 Detour to Convolutions......................................