Poisson Versus Negative Binomial Regression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Introduction to Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A
NESUG 2010 Statistics and Analysis An Animated Guide: An Introduction To Poisson Regression Russ Lavery, K&L Consulting Services, King of Prussia, PA, U.S.A. ABSTRACT: This paper will be a brief introduction to Poisson regression (theory, steps to be followed, complications and interpretation) via a worked example. It is hoped that this will increase motivation towards learning this useful statistical technique. INTRODUCTION: Poisson regression is available in SAS through the GENMOD procedure (generalized modeling). It is appropriate when: 1) the process that generates the conditional Y distributions would, theoretically, be expected to be a Poisson random process and 2) when there is no evidence of overdispersion and 3) when the mean of the marginal distribution is less than ten (preferably less than five and ideally close to one). THE POISSON DISTRIBUTION: The Poison distribution is a discrete Percent of observations where the random variable X is expected distribution and is appropriate for to have the value x, given that the Poisson distribution has a mean modeling counts of observations. of λ= P(X=x, λ ) = (e - λ * λ X) / X! Counts are observed cases, like the 0.4 count of measles cases in cities. You λ can simply model counts if all data 0.35 were collected in the same measuring 0.3 unit (e.g. the same number of days or 0.3 0.5 0.8 same number of square feet). 0.25 λ 1 0.2 3 You can use the Poisson Distribution = 5 for modeling rates (rates are counts 0.15 20 per unit) if the units of collection were 8 different. -

Effect of Probability Distribution of the Response Variable in Optimal Experimental Design with Applications in Medicine †
mathematics Article Effect of Probability Distribution of the Response Variable in Optimal Experimental Design with Applications in Medicine † Sergio Pozuelo-Campos *,‡ , Víctor Casero-Alonso ‡ and Mariano Amo-Salas ‡ Department of Mathematics, University of Castilla-La Mancha, 13071 Ciudad Real, Spain; [email protected] (V.C.-A.); [email protected] (M.A.-S.) * Correspondence: [email protected] † This paper is an extended version of a published conference paper as a part of the proceedings of the 35th International Workshop on Statistical Modeling (IWSM), Bilbao, Spain, 19–24 July 2020. ‡ These authors contributed equally to this work. Abstract: In optimal experimental design theory it is usually assumed that the response variable follows a normal distribution with constant variance. However, some works assume other probability distributions based on additional information or practitioner’s prior experience. The main goal of this paper is to study the effect, in terms of efficiency, when misspecification in the probability distribution of the response variable occurs. The elemental information matrix, which includes information on the probability distribution of the response variable, provides a generalized Fisher information matrix. This study is performed from a practical perspective, comparing a normal distribution with the Poisson or gamma distribution. First, analytical results are obtained, including results for the linear quadratic model, and these are applied to some real illustrative examples. The nonlinear 4-parameter Hill model is next considered to study the influence of misspecification in a Citation: Pozuelo-Campos, S.; dose-response model. This analysis shows the behavior of the efficiency of the designs obtained in Casero-Alonso, V.; Amo-Salas, M. -

Lecture.7 Poisson Distributions - Properties, Normal Distributions- Properties
Lecture.7 Poisson Distributions - properties, Normal Distributions- properties Theoretical Distributions Theoretical distributions are 1. Binomial distribution Discrete distribution 2. Poisson distribution 3. Normal distribution Continuous distribution Discrete Probability distribution Bernoulli distribution A random variable x takes two values 0 and 1, with probabilities q and p ie., p(x=1) = p and p(x=0)=q, q-1-p is called a Bernoulli variate and is said to be Bernoulli distribution where p and q are probability of success and failure. It was given by Swiss mathematician James Bernoulli (1654-1705) Example • Tossing a coin(head or tail) • Germination of seed(germinate or not) Binomial distribution Binomial distribution was discovered by James Bernoulli (1654-1705). Let a random experiment be performed repeatedly and the occurrence of an event in a trial be called as success and its non-occurrence is failure. Consider a set of n independent trails (n being finite), in which the probability p of success in any trail is constant for each trial. Then q=1-p is the probability of failure in any trail. 1 The probability of x success and consequently n-x failures in n independent trails. But x successes in n trails can occur in ncx ways. Probability for each of these ways is pxqn-x. P(sss…ff…fsf…f)=p(s)p(s)….p(f)p(f)…. = p,p…q,q… = (p,p…p)(q,q…q) (x times) (n-x times) Hence the probability of x success in n trials is given by x n-x ncx p q Definition A random variable x is said to follow binomial distribution if it assumes non- negative values and its probability mass function is given by P(X=x) =p(x) = x n-x ncx p q , x=0,1,2…n q=1-p 0, otherwise The two independent constants n and p in the distribution are known as the parameters of the distribution. -

1 One Parameter Exponential Families
1 One parameter exponential families The world of exponential families bridges the gap between the Gaussian family and general dis- tributions. Many properties of Gaussians carry through to exponential families in a fairly precise sense. • In the Gaussian world, there exact small sample distributional results (i.e. t, F , χ2). • In the exponential family world, there are approximate distributional results (i.e. deviance tests). • In the general setting, we can only appeal to asymptotics. A one-parameter exponential family, F is a one-parameter family of distributions of the form Pη(dx) = exp (η · t(x) − Λ(η)) P0(dx) for some probability measure P0. The parameter η is called the natural or canonical parameter and the function Λ is called the cumulant generating function, and is simply the normalization needed to make dPη fη(x) = (x) = exp (η · t(x) − Λ(η)) dP0 a proper probability density. The random variable t(X) is the sufficient statistic of the exponential family. Note that P0 does not have to be a distribution on R, but these are of course the simplest examples. 1.0.1 A first example: Gaussian with linear sufficient statistic Consider the standard normal distribution Z e−z2=2 P0(A) = p dz A 2π and let t(x) = x. Then, the exponential family is eη·x−x2=2 Pη(dx) / p 2π and we see that Λ(η) = η2=2: eta= np.linspace(-2,2,101) CGF= eta**2/2. plt.plot(eta, CGF) A= plt.gca() A.set_xlabel(r'$\eta$', size=20) A.set_ylabel(r'$\Lambda(\eta)$', size=20) f= plt.gcf() 1 Thus, the exponential family in this setting is the collection F = fN(η; 1) : η 2 Rg : d 1.0.2 Normal with quadratic sufficient statistic on R d As a second example, take P0 = N(0;Id×d), i.e. -

ONE SAMPLE TESTS the Following Data Represent the Change
1 WORKED EXAMPLES 6 INTRODUCTION TO STATISTICAL METHODS EXAMPLE 1: ONE SAMPLE TESTS The following data represent the change (in ml) in the amount of Carbon monoxide transfer (an indicator of improved lung function) in smokers with chickenpox over a one week period: 33, 2, 24, 17, 4, 1, -6 Is there evidence of significant improvement in lung function (a) if the data are normally distributed with σ = 10, (b) if the data are normally distributed with σ unknown? Use a significance level of α = 0.05. SOLUTION: (a) Here we have a sample of size 7 with sample mean x = 10.71. We want to test H0 : μ = 0.0, H1 : μ = 0.0, 6 under the assumption that the data follow a Normal distribution with σ = 10.0 known. Then, we have, in the Z-test, 10.71 0.0 z = − = 2.83, 10.0/√7 which lies in the critical region, as the critical values for this test are 1.96, for significance ± level α = 0.05. Therefore we have evidence to reject H0. The p-value is given by p = 2Φ( 2.83) = 0.004 < α. − (b) The sample variance is s2 = 14.192. In the T-test, we have test statistic t given by x 0.0 10.71 0.0 t = − = − = 2.00. s/√n 14.19/√7 The upper critical value CR is obtained by solving FSt(n 1)(CR) = 0.975, − where FSt(n 1) is the cdf of a Student-t distribution with n 1 degrees of freedom; here n = 7, so − − we can use statistical tables or a computer to find that CR = 2.447, and note that, as Student-t distributions are symmetric the lower critical value is CR. -

Overdispersion, and How to Deal with It in R and JAGS (Requires R-Packages AER, Coda, Lme4, R2jags, Dharma/Devtools) Carsten F
Overdispersion, and how to deal with it in R and JAGS (requires R-packages AER, coda, lme4, R2jags, DHARMa/devtools) Carsten F. Dormann 07 December, 2016 Contents 1 Introduction: what is overdispersion? 1 2 Recognising (and testing for) overdispersion 1 3 “Fixing” overdispersion 5 3.1 Quasi-families . .5 3.2 Different distribution (here: negative binomial) . .6 3.3 Observation-level random effects (OLRE) . .8 4 Overdispersion in JAGS 12 1 Introduction: what is overdispersion? Overdispersion describes the observation that variation is higher than would be expected. Some distributions do not have a parameter to fit variability of the observation. For example, the normal distribution does that through the parameter σ (i.e. the standard deviation of the model), which is constant in a typical regression. In contrast, the Poisson distribution has no such parameter, and in fact the variance increases with the mean (i.e. the variance and the mean have the same value). In this latter case, for an expected value of E(y) = 5, we also expect that the variance of observed data points is 5. But what if it is not? What if the observed variance is much higher, i.e. if the data are overdispersed? (Note that it could also be lower, underdispersed. This is less often the case, and not all approaches below allow for modelling underdispersion, but some do.) Overdispersion arises in different ways, most commonly through “clumping”. Imagine the number of seedlings in a forest plot. Depending on the distance to the source tree, there may be many (hundreds) or none. The same goes for shooting stars: either the sky is empty, or littered with shooting stars. -

6.1 Definition: the Density Function of Th
CHAPTER 6: Some Continuous Probability Distributions Continuous Uniform Distribution: 6.1 Definition: The density function of the continuous random variable X on the interval [A; B] is 1 A x B B A ≤ ≤ f(x; A; B) = 8 − < 0 otherwise: : Application: Some continuous random variables in the physical, management, and biological sciences have approximately uniform probability distributions. For example, suppose we are counting events that have a Poisson distribution, such as telephone calls coming into a switchboard. If it is known that exactly one such event has occurred in a given interval, say (0; t),then the actual time of occurrence is distributed uniformly over this interval. Example: Arrivals of customers at a certain checkout counter follow a Poisson distribution. It is known that, during a given 30-minute period, one customer arrived at the counter. Find the probability that the customer arrived during the last 5 minutes of the 30-minute period. Solution: As just mentioned, the actual time of arrival follows a uniform distribution over the interval of (0; 30). If X denotes the arrival time, then 30 1 30 25 1 P (25 X 30) = dx = − = ≤ ≤ Z25 30 30 6 Theorem 6.1: The mean and variance of the uniform distribution are 2 B 2 2 B 1 x B A A+B µ = A x B A = 2(B A) = 2(B− A) = 2 : R − h − iA − It is easy to show that (B A)2 σ2 = − 12 Normal Distribution: 6.2 Definition: The density function of the normal random variable X, with mean µ and variance σ2, is 2 1 (1=2)[(x µ)/σ] n(x; µ, σ) = e− − < x < ; p2πσ − 1 1 where π = 3:14159 : : : and e = 2:71828 : : : Example: The SAT aptitude examinations in English and Mathematics were originally designed so that scores would be approximately normal with µ = 500 and σ = 100. -
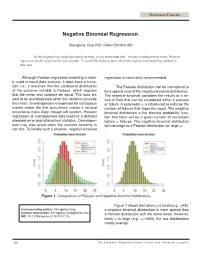
Negative Binomial Regression
STATISTICS COLUMN Negative Binomial Regression Shengping Yang PhD ,Gilbert Berdine MD In the hospital stay study discussed recently, it was mentioned that “in case overdispersion exists, Poisson regression model might not be appropriate.” I would like to know more about the appropriate modeling method in that case. Although Poisson regression modeling is wide- regression is commonly recommended. ly used in count data analysis, it does have a limita- tion, i.e., it assumes that the conditional distribution The Poisson distribution can be considered to of the outcome variable is Poisson, which requires be a special case of the negative binomial distribution. that the mean and variance be equal. The data are The negative binomial considers the results of a se- said to be overdispersed when the variance exceeds ries of trials that can be considered either a success the mean. Overdispersion is expected for contagious or failure. A parameter ψ is introduced to indicate the events where the first occurrence makes a second number of failures that stops the count. The negative occurrence more likely, though still random. Poisson binomial distribution is the discrete probability func- regression of overdispersed data leads to a deflated tion that there will be a given number of successes standard error and inflated test statistics. Overdisper- before ψ failures. The negative binomial distribution sion may also result when the success outcome is will converge to a Poisson distribution for large ψ. not rare. To handle such a situation, negative binomial 0 5 10 15 20 25 0 5 10 15 20 25 Figure 1. Comparison of Poisson and negative binomial distributions. -

Statistical Inference
GU4204: Statistical Inference Bodhisattva Sen Columbia University February 27, 2020 Contents 1 Introduction5 1.1 Statistical Inference: Motivation.....................5 1.2 Recap: Some results from probability..................5 1.3 Back to Example 1.1...........................8 1.4 Delta method...............................8 1.5 Back to Example 1.1........................... 10 2 Statistical Inference: Estimation 11 2.1 Statistical model............................. 11 2.2 Method of Moments estimators..................... 13 3 Method of Maximum Likelihood 16 3.1 Properties of MLEs............................ 20 3.1.1 Invariance............................. 20 3.1.2 Consistency............................ 21 3.2 Computational methods for approximating MLEs........... 21 3.2.1 Newton's Method......................... 21 3.2.2 The EM Algorithm........................ 22 1 4 Principles of estimation 23 4.1 Mean squared error............................ 24 4.2 Comparing estimators.......................... 25 4.3 Unbiased estimators........................... 26 4.4 Sufficient Statistics............................ 28 5 Bayesian paradigm 33 5.1 Prior distribution............................. 33 5.2 Posterior distribution........................... 34 5.3 Bayes Estimators............................. 36 5.4 Sampling from a normal distribution.................. 37 6 The sampling distribution of a statistic 39 6.1 The gamma and the χ2 distributions.................. 39 6.1.1 The gamma distribution..................... 39 6.1.2 The Chi-squared distribution.................. 41 6.2 Sampling from a normal population................... 42 6.3 The t-distribution............................. 45 7 Confidence intervals 46 8 The (Cramer-Rao) Information Inequality 51 9 Large Sample Properties of the MLE 57 10 Hypothesis Testing 61 10.1 Principles of Hypothesis Testing..................... 61 10.2 Critical regions and test statistics.................... 62 10.3 Power function and types of error.................... 64 10.4 Significance level............................ -

Generalized Linear Models
CHAPTER 6 Generalized linear models 6.1 Introduction Generalized linear modeling is a framework for statistical analysis that includes linear and logistic regression as special cases. Linear regression directly predicts continuous data y from a linear predictor Xβ = β0 + X1β1 + + Xkβk.Logistic regression predicts Pr(y =1)forbinarydatafromalinearpredictorwithaninverse-··· logit transformation. A generalized linear model involves: 1. A data vector y =(y1,...,yn) 2. Predictors X and coefficients β,formingalinearpredictorXβ 1 3. A link function g,yieldingavectoroftransformeddataˆy = g− (Xβ)thatare used to model the data 4. A data distribution, p(y yˆ) | 5. Possibly other parameters, such as variances, overdispersions, and cutpoints, involved in the predictors, link function, and data distribution. The options in a generalized linear model are the transformation g and the data distribution p. In linear regression,thetransformationistheidentity(thatis,g(u) u)and • the data distribution is normal, with standard deviation σ estimated from≡ data. 1 1 In logistic regression,thetransformationistheinverse-logit,g− (u)=logit− (u) • (see Figure 5.2a on page 80) and the data distribution is defined by the proba- bility for binary data: Pr(y =1)=y ˆ. This chapter discusses several other classes of generalized linear model, which we list here for convenience: The Poisson model (Section 6.2) is used for count data; that is, where each • data point yi can equal 0, 1, 2, ....Theusualtransformationg used here is the logarithmic, so that g(u)=exp(u)transformsacontinuouslinearpredictorXiβ to a positivey ˆi.ThedatadistributionisPoisson. It is usually a good idea to add a parameter to this model to capture overdis- persion,thatis,variationinthedatabeyondwhatwouldbepredictedfromthe Poisson distribution alone. -

Likelihood-Based Inference
The score statistic Inference Exponential distribution example Likelihood-based inference Patrick Breheny September 29 Patrick Breheny Survival Data Analysis (BIOS 7210) 1/32 The score statistic Univariate Inference Multivariate Exponential distribution example Introduction In previous lectures, we constructed and plotted likelihoods and used them informally to comment on likely values of parameters Our goal for today is to make this more rigorous, in terms of quantifying coverage and type I error rates for various likelihood-based approaches to inference With the exception of extremely simple cases such as the two-sample exponential model, exact derivation of these quantities is typically unattainable for survival models, and we must rely on asymptotic likelihood arguments Patrick Breheny Survival Data Analysis (BIOS 7210) 2/32 The score statistic Univariate Inference Multivariate Exponential distribution example The score statistic Likelihoods are typically easier to work with on the log scale (where products become sums); furthermore, since it is only relative comparisons that matter with likelihoods, it is more meaningful to work with derivatives than the likelihood itself Thus, we often work with the derivative of the log-likelihood, which is known as the score, and often denoted U: d U (θ) = `(θjX) X dθ Note that U is a random variable, as it depends on X U is a function of θ For independent observations, the score of the entire sample is the sum of the scores for the individual observations: X U = Ui i Patrick Breheny Survival -

(Introduction to Probability at an Advanced Level) - All Lecture Notes
Fall 2018 Statistics 201A (Introduction to Probability at an advanced level) - All Lecture Notes Aditya Guntuboyina August 15, 2020 Contents 0.1 Sample spaces, Events, Probability.................................5 0.2 Conditional Probability and Independence.............................6 0.3 Random Variables..........................................7 1 Random Variables, Expectation and Variance8 1.1 Expectations of Random Variables.................................9 1.2 Variance................................................ 10 2 Independence of Random Variables 11 3 Common Distributions 11 3.1 Ber(p) Distribution......................................... 11 3.2 Bin(n; p) Distribution........................................ 11 3.3 Poisson Distribution......................................... 12 4 Covariance, Correlation and Regression 14 5 Correlation and Regression 16 6 Back to Common Distributions 16 6.1 Geometric Distribution........................................ 16 6.2 Negative Binomial Distribution................................... 17 7 Continuous Distributions 17 7.1 Normal or Gaussian Distribution.................................. 17 1 7.2 Uniform Distribution......................................... 18 7.3 The Exponential Density...................................... 18 7.4 The Gamma Density......................................... 18 8 Variable Transformations 19 9 Distribution Functions and the Quantile Transform 20 10 Joint Densities 22 11 Joint Densities under Transformations 23 11.1 Detour to Convolutions......................................