Profile of Philip Hieter PROFILE
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2004 Albert Lasker Nomination Form
albert and mary lasker foundation 110 East 42nd Street Suite 1300 New York, ny 10017 November 3, 2003 tel 212 286-0222 fax 212 286-0924 Greetings: www.laskerfoundation.org james w. fordyce On behalf of the Albert and Mary Lasker Foundation, I invite you to submit a nomination Chairman neen hunt, ed.d. for the 2004 Albert Lasker Medical Research Awards. President mrs. anne b. fordyce The Awards will be offered in three categories: Basic Medical Research, Clinical Medical Vice President Research, and Special Achievement in Medical Science. This is the 59th year of these christopher w. brody Treasurer awards. Since the program was first established in 1944, 68 Lasker Laureates have later w. michael brown Secretary won Nobel Prizes. Additional information on previous Lasker Laureates can be found jordan u. gutterman, m.d. online at our web site http://www.laskerfoundation.org. Representative Albert Lasker Medical Research Awards Program Nominations that have been made in previous years may be updated and resubmitted in purnell w. choppin, m.d. accordance with the instructions on page 2 of this nomination booklet. daniel e. koshland, jr., ph.d. mrs. william mccormick blair, jr. the honorable mark o. hatfied Nominations should be received by the Foundation no later than February 2, 2004. Directors Emeritus A distinguished panel of jurors will select the scientists to be honored. The 2004 Albert Lasker Medical Research Awards will be presented at a luncheon ceremony given by the Foundation in New York City on Friday, October 1, 2004. Sincerely, Joseph L. Goldstein, M.D. Chairman, Awards Jury Albert Lasker Medical Research Awards ALBERT LASKER MEDICAL2004 RESEARCH AWARDS PURPOSE AND DESCRIPTION OF THE AWARDS The major purpose of these Awards is to recognize and honor individuals who have made signifi- cant contributions in basic or clinical research in diseases that are the main cause of death and disability. -

Lasker Interactive Research Nom'18.Indd
THE 2018 LASKER MEDICAL RESEARCH AWARDS Nomination Packet albert and mary lasker foundation November 1, 2017 Greetings: On behalf of the Albert and Mary Lasker Foundation, I invite you to submit a nomination for the 2018 Lasker Medical Research Awards. Since 1945, the Lasker Awards have recognized the contributions of scientists, physicians, and public citizens who have made major advances in the understanding, diagnosis, treatment, cure, and prevention of disease. The Medical Research Awards will be offered in three categories in 2018: Basic Research, Clinical Research, and Special Achievement. The Lasker Foundation seeks nominations of outstanding scientists; nominations of women and minorities are encouraged. Nominations that have been made in previous years are not automatically reconsidered. Please see the Nomination Requirements section of this booklet for instructions on updating and resubmitting a nomination. The Foundation accepts electronic submissions. For information on submitting an electronic nomination, please visit www.laskerfoundation.org. Lasker Awards often presage future recognition of the Nobel committee, and they have become known popularly as “America’s Nobels.” Eighty-seven Lasker laureates have received the Nobel Prize, including 40 in the last three decades. Additional information on the Awards Program and on Lasker laureates can be found on our website, www.laskerfoundation.org. A distinguished panel of jurors will select the scientists to be honored with Lasker Medical Research Awards. The 2018 Awards will -
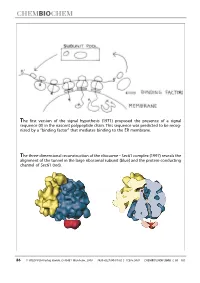
The First Version of the Signal Hypothesis (1971) Proposed the Presence of a Signal Sequence (X) in the Nascent Polypeptide Chain
The first version of the signal hypothesis (1971) proposed the presence of a signal sequence (X) in the nascent polypeptide chain. This sequence was predicted to be recog- nized by a ªbinding factorº that mediates binding to the ER membrane. The three-dimensional reconstruction of the ribosome ± Sec61 complex (1997) reveals the alignment of the tunnel in the large ribosomal subunit (blue) and the protein-conducting channel of Sec61 (red). 86 WILEY-VCH-Verlag GmbH, D-69451 Weinheim, 2000 1439-4227/00/01/02 $ 17.50+.50/0 CHEMBIOCHEM 2000,1,86±102 Protein Targeting (Nobel Lecture)** Günter Blobel*[a] KEYWORDS: membranes ´ Nobel lecture ´ proteins ´ protein transport Prologue I began research in the sixties, first as a graduate student in the laboratory of Van R. Potter (Figure 1) at the McArdle Institute for Cancer Research of the University of Wisconsin in Madison. I continued as a postdoctoral fellow in the laboratory of George E. Palade (Figure 2) at The Rockefeller Uni- versity in New York City. At that time, the intracellular pathway of secretory pro- teins, from their synthesis to their extru- sion from the cell, the so-called ªsecre- tory pathwayº, had already been worked out by George Palade and his co-workers, primarily Philip Siekevitz, Jim Jamieson, and Lucien Caro (for review see the 1974 Nobel lecture by George Palade[1] ). Using Figure 3. The secretory pathway. Secretory proteins (indicated in red) are pulse ± chase labeling with radioactive synthesized on ribosomes bound to the endoplasmic reticulum (ER). They are then amino acids in tissue slices in conjunction transported via vesicular carriers through the Golgi complex and are finally exocytosed. -

Cold Spring Harbor Laboratory of Quantitative
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Cold Spring Harbor Laboratory Institutional Repository COLD SPRING HARBOR LABORATORY OF QUANTITATIVE BIOLOGY COVER PHOTO: SAND SPIT AT COLD SPRING HARBOR COLD SPRING HARBOR LABORATORY OF QUANTITATIVE BIOLOGY ESTABLISHED JULY 1, 1963 FIRST ANNUAL REPORT CALENDAR YEAR 1964 COLD SPRING HARBOR, LONG ISLAND, NEW YORK COLD SPRING HARBOR LABORATORY OF QUANTITATIVE BIOLOGY Cold Spring Harbor, New York OFFICERS OF THE CORPORATION Chairman: Dr. Edward L. Tatum 1st Vice-chairman: Dr. Harry Eagle Laboratory Director: Dr. John Cairns 2nd Vice-chairman: Mr. Walter H. Page Secretary-Treasurer: Mr. John B. Philips BOARD OF TRUSTEES AND PARTICIPATING INSTITUTIONS The Rockefeller Institute Albert Einstein College of Medicine Dr. Edward L. Tatum Dr. Harry Eagle Long Island Biological Association Mr. Walter H. Page Duke University Dr. Philip Handler Princeton University Dr. Arthur B. Pardee Brooklyn College of the City University New York University Medical Center of New York of New York University Dr. Harry G. Albaum Dr. Alan W. Bernheimer Sloan-Kettering Institute for The Public Health Research Institute Cancer Research of the City of New York, Inc. Dr. Leo Wade Dr. George K. Hirst INDIVIDUAL TRUSTEES Mr. Robert V. Lindsay Mr. Dudley W. Stoddard TERMS OF OFFICE To expire 1965 To expire 1966 To expire 1967 Dr. Harry G. Albaum Dr. George K. Hirst Dr. Alan W. Bernheimer Dr. Philip Handler Mr. Robert V. Lindsay Dr. Harry Eagle Dr. Arthur B. Pardee Dr. Edward L. Tatum -

Harold E. Varmus
RETROVIRUSES AND ONCOGENES I Nobel Lecture, December 8, 1989 by HAROLD E. VARMUS Departments of Microbiology and Immunology, and Biochemistry and Biophysics, University of California at San Francisco, San Francisco, U.S.A. INTRODUCTION The story that Mike Bishop and I will tell in these two lectures is one in which retroviruses, oncogenes, our personal histories, and the history of tumor virology are closely interwoven. It begins with some simple questions about the origin and behavior of viral genes and takes us to a vantage point from which we can survey many aspects of retroviruses and animal cells, including some of the aberrations that lead to cancer. We now know that retroviruses capture normal cellular genes and convert them to cancer- causing genes, called oncogenes. Such transductions are rare, but depend upon the normal events of an intricate virus life cycle. Retroviruses have introduced us to more than forty cellular genes with the potential to become oncogenes, some discovered as components of viral genomes, others as genetic targets for viral insertion mutations. It has been our privilege to participate in a generous share of the experiments that established these principles. But we have required the help of many talented people in our laboratories at UCSF, as well as the collaboration and friendly competition of others elsewhere. (I mention as many names as the narrative can bear, but inevitably I must apologize to valued colleagues who remain anonymous here.) Several viruses also figure in our tale, but Rous sarcoma virus again has the leading role, yet another of many tributes to the pioneering work of Peyton Rous and to the principle of delayed gratification in science. -

Available As
Royal Netherlands Academy of Arts and Sciences Heineken Lectures 2004 Dr A.H. Heineken Prize for Environmental Sciences Simon A.Levin Heineken Lectures 2004 Dr A.H. Heineken Prize for Environmental Sciences Professor Simon A.Levin delivered his Heineken Lecture on September 28,2004 at the Royal Tropical Institute in Amsterdam. Royal Netherlands Academy of Arts and Sciences Heineken Lectures 2004 Amsterdam,2005 Dr A.H. Heineken Prize for Environmental Sciences Simon A.Levin The Heineken Lectures series for 2004 also includes Dr H.P.Heineken Prize for Biochemistry and Biophysics 2004,Andrew Z. Fire Dr A.H. Heineken Prize for Medicine 2004, Elizabeth H. Blackburn Dr A.H. Heineken Prize for History 2004, Jacques Le Goff To order publications from the Heineken Lectures series for 2004, please contact the secretariat of the Academy’s Communications Department at +31 (0)20 551 0726. Contents page 6 Preface Willem J.M.Levelt 9 Simon A. Levin and his research 12 Presentation address for the Dr A.H. Heineken Prize for Environmental Sciences Johan Bouma 14 The Ecology of Complexity and the Complexity of Ecology Dr A.H. Heineken Prize for Environmental Sciences Simon A.Levin 26 Heineken Lecture Program 2004 27 Audience and publicity for the Heineken Lectures in 2004 28 Secondary School Project about the Heineken laureates 29 General information 30 List of Heineken laureates Preface Address by Willem Levelt,President of the Royal Netherlands Academy of Arts and Sciences,on the occasion of the presentation of the 2004 Heineken Prizes on October 1,2004. Both in the sciences and the arts, history provides us with a looking glass that helps us to focus on the real pioneers. -

Leadership Innovation Impact Annual Report 20 20
LEADERSHIP INNOVATION ANNUAL 20 IMPACT REPORT 20 TABLE OF CONTENTS 04 Letter from the President 06 2021-2022 Governing Council 08 Organizational Chart 09 Commitment to Racial Equity 11 Program Highlights 12 RESPONDING TO CRITICAL & PRESSING ISSUES 12 / Mobilizing to Counter COVID-19 14 / Confronting the U.S. Opioid Epidemic 16 / Championing Clinician Well-Being & Resilience 18 / Human Germline Genome Editing 20 / Climate Change & Human Health 22 / Preparing for Seasonal and Pandemic Influenza 23 ADVISING THE NATION & THE WORLD ON HEALTH & HEALTH CARE 23 / NAM Leadership Consortium 28 / Advancing Health Equity 30 / Vital Directions in Health & Health Care 32 / The Future of Nursing 33 LEADING & INSPIRING FOR THE FUTURE 33 / Healthy Longevity Global Grand Challenge 35 / Committee on Emerging Science, Technology, and Innovation (CESTI) 36 / Global Health Leadership 37 / NAM Perspectives 39 Member Highlights 40 / Members Elected in 2019 43 / Members Elected in 2020 46 / 2020 Nobel Laureates 47 / 2020 Annual Meeting 49 / The IOM/NAM Anniversary Celebration 50 / In Memoriam 51 Fellowships & Leadership Programs 57 Awards 62 Finances 64 Donor Appreciation 75 Contact Us LETTER FROM THE PRESIDENT Responding to COVID-19 At the outset of 2020, the NAM was preparing to celebrate the 50th anniversary of the founding of the Institute of Medicine (IOM) and the 5th anniversary of IOM’s reconstitution as the NAM. Then, the COVID-19 global pandemic struck. By June 2021, the virus had claimed about 3.5 million lives worldwide and infected over 170 million people. Beyond creating an unprecedented health crisis, the pandemic has threatened the global economy, political stability, and social structure. -

Top 100 Living Contributors to Biotechnology
Over the last 30 years, a small group of visionaries in science, technology, legislation and business have driven the development of biotechnology. THE Today, in the midst of tremendous advances in medicine and agriculture, this exhibition and accompanying brochure pays tribute to the leaders that have shaped the biotechnology industry. TOP The Top 100 Living Contributors to Biotechnology have been selected by their peers and through independent polls conducted by Reed Exhibitions, a division of Reed Elsevier. Senior staff throughout the biotechnology industry have identified the most influential and inspirational pioneers. The results 100 are presented here alphabetically. LIVING CONTRIBUTORS To those named in the Top 100, and the many other contributors not listed, TO BIOTECHNOLOGY the biotechnology community is deeply appreciative. P 1 4 MICHAEL ASHBURNER SEYMOUR BENZER PAUL BERG Michael Ashburner is Professor Seymour Benzer instilled the Paul Berg is Cahill Professor in of Biology at the University of fundamental idea that genes Cancer Research, Emeritus, at Cambridge where he received his control behaviour. He began his the Stanford University School undergraduate degree and PhD, career studying gene structure of Medicine, and director emeri- both in genetics. Ashburner’s and code, developing a method tus of the Beckman Centre for current major research interests to determine the detailed struc- Molecular and Genetic are the structure and evolution of ture of viral genes in 1955. He Medicine. He is one of the prin- genomes. Most of his research then switched to the field of cipal pioneers in the field of 33 has been with the model organ- neurogenetics, focusing on “gene splicing.” Berg, along with ism Drosophila melanogaster, the inheritance of behaviour. -
Molecular Biology of Cancer: Mechanisms, Targets
Molecular Biology of Cancer This page intentionally left blank Molecular Biology of Cancer Mechanisms, Targets, and Therapeutics THIRD EDITION Lauren Pecorino University of Greenwich 1 1 Great Clarendon Street, Oxford, OX2 6DP, United Kingdom Oxford University Press is a department of the University of Oxford. It furthers the University’s objective of excellence in research, scholarship, and education by publishing worldwide. Oxford is a registered trade mark of Oxford University Press in the UK and in certain other countries © Lauren Pecorino 2012 The moral rights of the author have been asserted First Edition copyright 2005 Second Edition copyright 2008 Impression: 1 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, without the prior permission in writing of Oxford University Press, or as expressly permitted by law, by licence or under terms agreed with the appropriate reprographics rights organization. Enquiries concerning reproduction outside the scope of the above should be sent to the Rights Department, Oxford University Press, at the address above You must not circulate this work in any other form and you must impose this same condition on any acquirer British Library Cataloguing in Publication Data Data available Library of Congress Cataloging in Publication Data Library of Congress Control Number: 2012932747 ISBN 978–0–19–957717–0 Printed in Italy on acid-free paper by L.E.G.O. S.p.A.—Lavis TN Links to third party websites are provided by Oxford in good faith and for information only. Oxford disclaims any responsibility for the materials contained in any third party website referenced in this work. -

Proceedings National Academy of Sciences
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA October 1, 1996 Volume 93, Number 20 pp. 10539–11286 Table of Contents AUTHOR INDEX xi–xiii INFORMATION FOR CONTRIBUTORS xiv–xv COPYRIGHT ASSIGNMENT FORM xvi Perspective Physical Sciences How Charles Nicolle of the Pasteur Institute 10539–10540 discovered that epidemic typhus is transmitted by lice: Reminiscences from my years at the APPLIED PHYSICAL SCIENCES Pasteur Institute in Paris Ludwik Gross Room-temperature single-electron junction 10556–10559 Paolo Facci, Victor Erokhin, Sandro Carrara, and Claudio Nicolini Commentaries The weaver mouse: A most cantankerous rodent 10541–10542 Karl Herrup CHEMISTRY What the nose knows: New understandings of 10543–10545 Neanderthal upper respiratory tract specializations Acyl tunichlorins: A new class of nickel chlorins 10560–10565 Jeffrey T. Laitman, Joy S. Reidenberg, isolated from the Caribbean tunicate Samuel Marquez, and Patrick J. Gannon Trididemnum solidum Heather L. Sings, Keith C. Bible, Learning and receptive field plasticity 10546–10547 Charles D. Gilbert and Kenneth L. Rinehart Cation-p interactions in aromatics of biological and 10566–10571 Review medicinal interest: Electrostatic potential surfaces as a useful qualitative guide What determines the folding of the chromatin fiber? 10548–10555 Sandro Mecozzi, Anthony P. West, Jr., and Dennis Kensal van Holde and Jordanka Zlatanova A. Dougherty iii Downloaded by guest on September 27, 2021 Contents Biological Sciences Mutations in the C-terminal fragment of DnaK 10632–10637 affecting peptide binding William F. Burkholder, Xun Zhao, Xiaotian Zhu, APPLIED BIOLOGICAL SCIENCES Wayne A. Hendrickson, Alexander Gragerov, Transcriptional regulation of the Rhodococcus 10572–10577 and Max E. -

ANNEX A: Highlights of the Biotechnology Revolution: 1953
Annex A: Highlights of the “Biotechnology Revolution”: 1953–present* 1953 Nature magazine published James Watson’s and Francis Crick’s manuscript which described the double helix structure of DNA. The discovery of the structure of DNA resulted in an explosion of research in molecular biology and genetics, paving the way for the “biotechnology revolution.” 1955 Seymour Benzer at Purdue University devised an experimental setup to map mutations within a short genetic region of a particular bacterial virus. Over a five-year period, Benzer mapped recombinations of genetic material that distinguished mutational changes that had taken place at adjacent base pairs. 1956 Heinz Fraenkel-Conrat took apart and reassembled the tobacco mosaic virus, demonstrating “self assembly.” 1957 Francis Crick and George Gamov worked out the “central dogma,” explaining how DNA functions to make protein. Their “sequence hypothesis” posited that the DNA sequence specifies the amino acid sequence in a protein. They also suggested that genetic information flows only in one direction, from DNA to messenger RNA to protein, the central concept of the central dogma. 1957 Matthew Meselson and Frank Stahl demonstrated the replication mechanism of DNA. 1958 Coenberg discovered and isolated DNA polymerase, which became the first enzyme used to make DNA in a test tube. 1958 The National Seed Storage Laboratory (NSSI) was opened in Fort Collins, Colorado, becoming the first long-term seed storage facility in the world. 1959 Francois Jacob and Jacques Monod established the existence of genetic regulation—mappable control functions located on the chromosome in the DNA sequence—which they named the repressor and operon. -

2013 Sonneborn Program Wieschaus.Indd
Indiana University Department of Biology Spring 2013 Sonneborn Lecture Hoosier native Eric Wieschaus is known for his groundbreaking work with the fruit fl y, Drosophila melanogaster. Wieschaus received the Nobel Prize in Physiology or Medicine with Christiane Nüsslein-Volhard and Edward B. Lewis for their discoveries concerning the genetic control of early embryonic development. These developmental biologists were able to identify and classify a small number of genes that are of key importance in determining the body plan and the formation of body segments. Wieschaus and Nüsslein-Volhard established that genes controlling development could be systematically identifi ed, that the number of genes involved was limited, and that they could be classifi ed into specifi c functional groups. This encouraged a number of other scientists to look for developmental genes in other species, and in a relatively short time it was possible to show that similar or identical genes also existed in higher organisms and in man. Three years after their research was published, in 1980 in the journal Nature, researchers Matthew Scott and Amy Weiner, working in the Indiana University Bloomington laboratory of Distinguished Professor of Biology Thomas Kaufman, identifi ed the homeobox in fruit fl ies, a DNA- binding gene sequence that encodes proteins, which tell cells what kinds of structures to make in various segments of a developing embryo. Shortly after that discovery homeobox genes were quickly found in the genomes of mice and humans. “Eric and his colleague Janni Nüsslein-Volhard bravely Photo courtesy of Princeton University, Offi ce of Communications embarked on a screen for mutations in Drosophila that affected embryonic patterning,” Kaufman said.