The Interrupted Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Discovery of DNA Structure and Function: Watson and Crick By: Leslie A
01/08/2018 Discovery of DNA Double Helix: Watson and Crick | Learn Science at Scitable NUCLEIC ACID STRUCTURE AND FUNCTION | Lead Editor: Bob Moss Discovery of DNA Structure and Function: Watson and Crick By: Leslie A. Pray, Ph.D. © 2008 Nature Education Citation: Pray, L. (2008) Discovery of DNA structure and function: Watson and Crick. Nature Education 1(1):100 The landmark ideas of Watson and Crick relied heavily on the work of other scientists. What did the duo actually discover? Aa Aa Aa Many people believe that American biologist James Watson and English physicist Francis Crick discovered DNA in the 1950s. In reality, this is not the case. Rather, DNA was first identified in the late 1860s by Swiss chemist Friedrich Miescher. Then, in the decades following Miescher's discovery, other scientists--notably, Phoebus Levene and Erwin Chargaff--carried out a series of research efforts that revealed additional details about the DNA molecule, including its primary chemical components and the ways in which they joined with one another. Without the scientific foundation provided by these pioneers, Watson and Crick may never have reached their groundbreaking conclusion of 1953: that the DNA molecule exists in the form of a three-dimensional double helix. The First Piece of the Puzzle: Miescher Discovers DNA Although few people realize it, 1869 was a landmark year in genetic research, because it was the year in which Swiss physiological chemist Friedrich Miescher first identified what he called "nuclein" inside the nuclei of human white blood cells. (The term "nuclein" was later changed to "nucleic acid" and eventually to "deoxyribonucleic acid," or "DNA.") Miescher's plan was to isolate and characterize not the nuclein (which nobody at that time realized existed) but instead the protein components of leukocytes (white blood cells). -

Biochemistrystanford00kornrich.Pdf
University of California Berkeley Regional Oral History Office University of California The Bancroft Library Berkeley, California Program in the History of the Biosciences and Biotechnology Arthur Kornberg, M.D. BIOCHEMISTRY AT STANFORD, BIOTECHNOLOGY AT DNAX With an Introduction by Joshua Lederberg Interviews Conducted by Sally Smith Hughes, Ph.D. in 1997 Copyright 1998 by The Regents of the University of California Since 1954 the Regional Oral History Office has been interviewing leading participants in or well-placed witnesses to major events in the development of Northern California, the West, and the Nation. Oral history is a method of collecting historical information through tape-recorded interviews between a narrator with firsthand knowledge of historically significant events and a well- informed interviewer, with the goal of preserving substantive additions to the historical record. The tape recording is transcribed, lightly edited for continuity and clarity, and reviewed by the interviewee. The corrected manuscript is indexed, bound with photographs and illustrative materials, and placed in The Bancroft Library at the University of California, Berkeley, and in other research collections for scholarly use. Because it is primary material, oral history is not intended to present the final, verified, or complete narrative of events. It is a spoken account, offered by the interviewee in response to questioning, and as such it is reflective, partisan, deeply involved, and irreplaceable. ************************************ All uses of this manuscript are covered by a legal agreement between The Regents of the University of California and Arthur Kornberg, M.D., dated June 18, 1997. The manuscript is thereby made available for research purposes. All literary rights in the manuscript, including the right to publish, are reserved to The Bancroft Library of the University of California, Berkeley. -

1 History for Canonical and Non-Canonical Structures of Nucleic Acids
1 1 History for Canonical and Non-canonical Structures of Nucleic Acids The main points of the learning: Understand canonical and non-canonical structures of nucleic acids and think of historical scientists in the research field of nucleic acids. 1.1 Introduction This book is to interpret the non-canonical structures and their stabilities of nucleic acids from the viewpoint of the chemistry and study their biological significances. There is more than 60 years’ history after the discovery of the double helix DNA structure by James Dewey Watson and Francis Harry Compton Crick in 1953, and chemical biology of nucleic acids is facing a new aspect today. Through this book, I expect that readers understand how the uncommon structure of nucleic acids became one of the common structures that fascinate us now. In this chapter, I introduce the history of nucleic acid structures and the perspective of research for non-canonical nucleic acid structures (see also Chapter 15). 1.2 History of Duplex The opening of the history of genetics was mainly done by three researchers. Charles Robert Darwin, who was a scientist of natural science, pioneered genetics. The proposition of genetic concept is indicated in his book On the Origin of Species published in 1859. He indicated the theory of biological evolution, which is the basic scientific hypothesis of natural diversity. In other words, he proposed biological evolution, which changed among individuals by adapting to the environment and be passed on to the next generation. However, that was still a primitive idea for the genetic concept. After that, Gregor Johann Mendel, who was a priest in Brno, Czech Republic, confirmed the mechanism of gene evolution by using “factor” inherited from parent to children using pea plant in 1865. -

2004 Albert Lasker Nomination Form
albert and mary lasker foundation 110 East 42nd Street Suite 1300 New York, ny 10017 November 3, 2003 tel 212 286-0222 fax 212 286-0924 Greetings: www.laskerfoundation.org james w. fordyce On behalf of the Albert and Mary Lasker Foundation, I invite you to submit a nomination Chairman neen hunt, ed.d. for the 2004 Albert Lasker Medical Research Awards. President mrs. anne b. fordyce The Awards will be offered in three categories: Basic Medical Research, Clinical Medical Vice President Research, and Special Achievement in Medical Science. This is the 59th year of these christopher w. brody Treasurer awards. Since the program was first established in 1944, 68 Lasker Laureates have later w. michael brown Secretary won Nobel Prizes. Additional information on previous Lasker Laureates can be found jordan u. gutterman, m.d. online at our web site http://www.laskerfoundation.org. Representative Albert Lasker Medical Research Awards Program Nominations that have been made in previous years may be updated and resubmitted in purnell w. choppin, m.d. accordance with the instructions on page 2 of this nomination booklet. daniel e. koshland, jr., ph.d. mrs. william mccormick blair, jr. the honorable mark o. hatfied Nominations should be received by the Foundation no later than February 2, 2004. Directors Emeritus A distinguished panel of jurors will select the scientists to be honored. The 2004 Albert Lasker Medical Research Awards will be presented at a luncheon ceremony given by the Foundation in New York City on Friday, October 1, 2004. Sincerely, Joseph L. Goldstein, M.D. Chairman, Awards Jury Albert Lasker Medical Research Awards ALBERT LASKER MEDICAL2004 RESEARCH AWARDS PURPOSE AND DESCRIPTION OF THE AWARDS The major purpose of these Awards is to recognize and honor individuals who have made signifi- cant contributions in basic or clinical research in diseases that are the main cause of death and disability. -

DNA: the Timeline and Evidence of Discovery
1/19/2017 DNA: The Timeline and Evidence of Discovery Interactive Click and Learn (Ann Brokaw Rocky River High School) Introduction For almost a century, many scientists paved the way to the ultimate discovery of DNA and its double helix structure. Without the work of these pioneering scientists, Watson and Crick may never have made their ground-breaking double helix model, published in 1953. The knowledge of how genetic material is stored and copied in this molecule gave rise to a new way of looking at and manipulating biological processes, called molecular biology. The breakthrough changed the face of biology and our lives forever. Watch The Double Helix short film (approximately 15 minutes) – hyperlinked here. 1 1/19/2017 1865 The Garden Pea 1865 The Garden Pea In 1865, Gregor Mendel established the foundation of genetics by unraveling the basic principles of heredity, though his work would not be recognized as “revolutionary” until after his death. By studying the common garden pea plant, Mendel demonstrated the inheritance of “discrete units” and introduced the idea that the inheritance of these units from generation to generation follows particular patterns. These patterns are now referred to as the “Laws of Mendelian Inheritance.” 2 1/19/2017 1869 The Isolation of “Nuclein” 1869 Isolated Nuclein Friedrich Miescher, a Swiss researcher, noticed an unknown precipitate in his work with white blood cells. Upon isolating the material, he noted that it resisted protein-digesting enzymes. Why is it important that the material was not digested by the enzymes? Further work led him to the discovery that the substance contained carbon, hydrogen, nitrogen and large amounts of phosphorus with no sulfur. -

Structure of DNA &
Structure Of DNA & RNA By Himanshu Dev VMMC & SJH DNA DNA Deoxyribonucleic acid DNA - a polymer of deoxyribo- nucleotides. Usually double stranded. And have double-helix structure. found in chromosomes, mitochondria and chloroplasts. It acts as the genetic material in most of the organisms. Carries the genetic information A Few Key Events Led to the Discovery of the Structure of DNA DNA as an acidic substance present in nucleus was first identified by Friedrich Meischer in 1868. He named it as ‘Nuclein’. Friedrich Meischer In 1953 , James Watson and Francis Crick, described a very simple but famous Double Helix model for the structure of DNA. FRANCIS CRICK AND JAMES WATSON The scientific framework for their breakthrough was provided by other scientists including Linus Pauling Rosalind Franklin and Maurice Wilkins Erwin Chargaff Rosalind Franklin She worked in same laboratory as Maurice Wilkins. She study X-ray diffraction to study wet fibers of DNA. X-ray diffraction of wet DNA fibers The diffraction pattern is interpreted X Ray (using mathematical theory) Crystallography This can ultimately provide Rosalind information concerning the structure Franklin’s photo of the molecule She made marked advances in X-ray diffraction techniques with DNA The diffraction pattern she obtained suggested several structural features of DNA Helical More than one strand 10 base pairs per complete turn Rosalind Franklin Maurice Wilkins DNA Structure DNA structure is often divided into four different levels primary, secondary, tertiary -
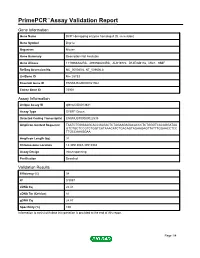
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DCP1 decapping enzyme homolog A (S. cerevisiae) Gene Symbol Dcp1a Organism Mouse Gene Summary Description Not Available Gene Aliases 1110066A22Rik, 4930568L04Rik, AU019772, D14Ertd817e, Mitc1, SMIF RefSeq Accession No. NC_000080.6, NT_039606.8 UniGene ID Mm.28733 Ensembl Gene ID ENSMUSG00000021962 Entrez Gene ID 75901 Assay Information Unique Assay ID qMmuCID0013841 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000022535 Amplicon Context Sequence TAATCTGGGAAGCACCGAGACTCTAGAAGAGACACCCTCTGGGTCACAGGATAA GTCTGCTCCGTCTGGTCATAAACATCTGACAGTAGAAGAGTTATTTGGAACCTCC TTGCCAAAGGAA Amplicon Length (bp) 91 Chromosome Location 14:30513043-30518984 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9997 cDNA Cq 22.41 cDNA Tm (Celsius) 81 gDNA Cq 24.87 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Dcp1a, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Exon Amplification: a Strategy to Isolate Mammalian Genes Based on RNA Splicing (Gene Cloning/Polymerase Chain Reaction) ALAN J
Proc. Natl. Acad. Sci. USA Vol. 88, pp. 4005-4009, May 1991 Genetics Exon amplification: A strategy to isolate mammalian genes based on RNA splicing (gene cloning/polymerase chain reaction) ALAN J. BUCKLER*, DAVID D. CHANG, SHARON L. GRAw, J. DAVID BROOK, DANIEL A. HABER, PHILLIP A. SHARP, AND DAVID E. HOUSMAN Center for Cancer Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139 Contributed by Phillip A. Sharp, January 25, 1991 ABSTRACT We have developed a method, exon amplifi- (7, 8). Thus, this method may be generally applicable for the cation, for fast and efficient isolation of coding sequences from selection of exon sequences from any gene. The method is complex mammalian genomic DNA. This method is based on also both rapid and easily adapted to large scale experiments. the selection of RNA sequences, exons, which are flanked by A series of cloned genomic DNA fragments can be screened functional 5' and 3' splice sites. Fragments of cloned genomic within 1-2 weeks. The sensitivity of this method is high. DNA are inserted into an intron, which is flanked by 5' and 3' Genomic DNA segments of 20 kilobases (kb) or more can be splice sites of the human immunodeficiency virus 1 tat gene successfully screened in a single transfection by using a set contained within the plasmid pSPL1. COS-7 cells are trans- of pooled subclones. This method thus allows the rapid fected with these constructs, and the resulting RNA transcripts identification of exons in mammalian genomic DNA and are processed in vivo. Splice sites of exons contained within the should facilitate the isolation of a wide spectrum of genes of inserted genomic fragment are paired with splice sites of the significance in physiology and development. -

Mrna Turnover Philip Mitchell* and David Tollervey†
320 mRNA turnover Philip Mitchell* and David Tollervey† Nuclear RNA-binding proteins can record pre-mRNA are cotransported to the cytoplasm with the mRNP. These processing events in the structure of messenger proteins may preserve a record of the nuclear history of the ribonucleoprotein particles (mRNPs). During initial rounds of pre-mRNA in the cytoplasmic mRNP structure. This infor- translation, the mature mRNP structure is established and is mation can strongly influence the cytoplasmic fate of the monitored by mRNA surveillance systems. Competition for the mRNA and is used by mRNA surveillance systems that act cap structure links translation and subsequent mRNA as a checkpoint of mRNP integrity, particularly in the identi- degradation, which may also involve multiple deadenylases. fication of premature translation termination codons (PTCs). Addresses Cotransport of nuclear mRNA-binding proteins with mRNA Wellcome Trust Centre for Cell Biology, ICMB, University of Edinburgh, from the nucleus to the cytoplasm (nucleocytoplasmic shut- Kings’ Buildings, Edinburgh EH9 3JR, UK tling) was first observed for the heterogeneous nuclear *e-mail: [email protected] ribonucleoprotein (hnRNP) proteins. Some hnRNP proteins †e-mail: [email protected] are stripped from the mRNA at export [1], but hnRNP A1, Current Opinion in Cell Biology 2001, 13:320–325 A2, E, I and K are all exported (see [2]). Although roles for 0955-0674/01/$ — see front matter these hnRNP proteins in transport and translation have been © 2001 Elsevier Science Ltd. All rights reserved. reported [3•,4•], their affects on mRNA stability have been little studied. More is known about hnRNP D/AUF1 and Abbreviations AREs AU-rich sequence elements another nuclear RNA-binding protein, HuR, which act CBC cap-binding complex antagonistically to modulate the stability of a range of DAN deadenylating nuclease mRNAs containing AU-rich sequence elements (AREs) DSEs downstream sequence elements (reviewed in [2]). -

Supplemental Information
Supplemental information Dissection of the genomic structure of the miR-183/96/182 gene. Previously, we showed that the miR-183/96/182 cluster is an intergenic miRNA cluster, located in a ~60-kb interval between the genes encoding nuclear respiratory factor-1 (Nrf1) and ubiquitin-conjugating enzyme E2H (Ube2h) on mouse chr6qA3.3 (1). To start to uncover the genomic structure of the miR- 183/96/182 gene, we first studied genomic features around miR-183/96/182 in the UCSC genome browser (http://genome.UCSC.edu/), and identified two CpG islands 3.4-6.5 kb 5’ of pre-miR-183, the most 5’ miRNA of the cluster (Fig. 1A; Fig. S1 and Seq. S1). A cDNA clone, AK044220, located at 3.2-4.6 kb 5’ to pre-miR-183, encompasses the second CpG island (Fig. 1A; Fig. S1). We hypothesized that this cDNA clone was derived from 5’ exon(s) of the primary transcript of the miR-183/96/182 gene, as CpG islands are often associated with promoters (2). Supporting this hypothesis, multiple expressed sequences detected by gene-trap clones, including clone D016D06 (3, 4), were co-localized with the cDNA clone AK044220 (Fig. 1A; Fig. S1). Clone D016D06, deposited by the German GeneTrap Consortium (GGTC) (http://tikus.gsf.de) (3, 4), was derived from insertion of a retroviral construct, rFlpROSAβgeo in 129S2 ES cells (Fig. 1A and C). The rFlpROSAβgeo construct carries a promoterless reporter gene, the β−geo cassette - an in-frame fusion of the β-galactosidase and neomycin resistance (Neor) gene (5), with a splicing acceptor (SA) immediately upstream, and a polyA signal downstream of the β−geo cassette (Fig. -

(Bsc Zoology and Microbiology) Concept of Introns and Exons
Unit-5 Molecular Biology (BSc Zoology and Microbiology) Concept of introns and exons Most of the portion of a gene in higher eukaryotes consists of noncoding DNA that interrupts the relatively short segments of coding DNA. The coding sequences are called exons. The noncoding sequences are called introns. Intron: An intron is a portion of a gene that does not code for amino acids An intron is any nucleotide sequence within a gene which is represented in the primary transcript of the gene, but not present in the final processed form. In other words, Introns are noncoding regions of an RNA transcript which are eliminated by splicing before translation. Sequences that are joined together in the final mature RNA after RNA splicing are exons. Introns are very large chunks of RNA within a messenger RNA molecule that interfere with the code of the exons. And these introns get removed from the RNA molecule to leave a string of exons attached to each other so that the appropriate amino acids can be encoded for. Introns are rare in genes of prokaryotes. #Look carefully at the diagram above, we have already discussed about the modification and processing of eukaryotic RNA. In which 5’ guanine cap and 3’poly A tail is added. So at that time, noncoding regions i.e. introns are removed. We hv done ths already. Ok Exon: The coding sequences are called Exon. An exon is the portion of a gene that codes for amino acids. In the cells of plants and animals, most gene sequences are broken up by one or more DNA sequences called introns. -

Lasker Interactive Research Nom'18.Indd
THE 2018 LASKER MEDICAL RESEARCH AWARDS Nomination Packet albert and mary lasker foundation November 1, 2017 Greetings: On behalf of the Albert and Mary Lasker Foundation, I invite you to submit a nomination for the 2018 Lasker Medical Research Awards. Since 1945, the Lasker Awards have recognized the contributions of scientists, physicians, and public citizens who have made major advances in the understanding, diagnosis, treatment, cure, and prevention of disease. The Medical Research Awards will be offered in three categories in 2018: Basic Research, Clinical Research, and Special Achievement. The Lasker Foundation seeks nominations of outstanding scientists; nominations of women and minorities are encouraged. Nominations that have been made in previous years are not automatically reconsidered. Please see the Nomination Requirements section of this booklet for instructions on updating and resubmitting a nomination. The Foundation accepts electronic submissions. For information on submitting an electronic nomination, please visit www.laskerfoundation.org. Lasker Awards often presage future recognition of the Nobel committee, and they have become known popularly as “America’s Nobels.” Eighty-seven Lasker laureates have received the Nobel Prize, including 40 in the last three decades. Additional information on the Awards Program and on Lasker laureates can be found on our website, www.laskerfoundation.org. A distinguished panel of jurors will select the scientists to be honored with Lasker Medical Research Awards. The 2018 Awards will