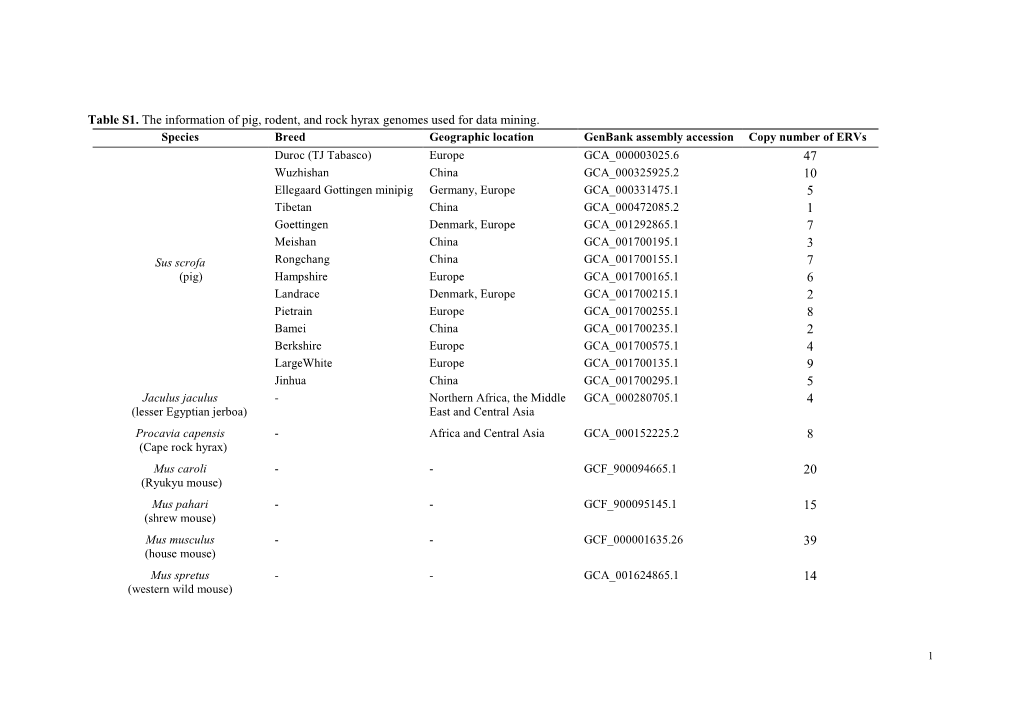
Table S1. the Information of Pig, Rodent, and Rock Hyrax Genomes Used for Data Mining
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Morphological Comparison of Ryukyu Mouse Mus Caroli
Zoological Studies 42(2): 258-267 (2003) Morphological Comparison of Ryukyu Mouse Mus caroli (Rodentia: Muridae) Populations from Okinawajima and Taiwan Masaharu Motokawa1,*, Liang-Kong Lin2 and Junko Motokawa3 1The Kyoto University Museum, Kyoto 606-8501, Japan 2Laboratory of Wildlife Ecology, Department of Biology, Tunghai University, Taichung, Taiwan 407, R.O.C. 3Department of Zoology, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan (Accepted January 14, 2003) Masaharu Motokawa, Liang-Kong Lin and Junko Motokawa (2003) Morphological comparison of Ryukyu mouse Mus caroli (Rodentia: Muridae) populations from Okinawajima and Taiwan. Zoological Studies 42(2): 258-267. We performed univariate, bivariate, and multivariate analyses of 4 external and 17 cranial morphome- tric characters in the Ryukyu mouse, Mus caroli Bonhote, 1902 (Mammalia: Rodentia: Muridae), using 177 specimens collected from Okinawajima in the central Ryukyus and from Taiwan. There were clear morphologi- cal differences between populations from Okinawajima and Taiwan. The univariate and bivariate analyses indi- cated that the Okinawajima population differs from the Taiwan population by a shorter tail, smaller ear and audi- tory bulla, less robust incisors, larger molar row, narrower cranium in the orbital region, and longer postpalatal region. Principal component and canonical discriminant analyses based on cranial variables also suggested morphological divergence between the 2 populations. http://www.sinica.edu.tw/zool/zoolstud/42.2/258.pdf Key words: Mus caroli, M. formosanus, Taxonomy, Morphometric analyses, Allometry. The Ryukyu mouse, Mus caroli, is distrib- ognize it as a valid species (e.g., Lin and Lin uted in the Ryukyu Archipelago (Japan), Taiwan, 1983). Hainan, and southern China to the Malay Another name included in M. -

Report on Biodiversity and Tropical Forests in Indonesia
Report on Biodiversity and Tropical Forests in Indonesia Submitted in accordance with Foreign Assistance Act Sections 118/119 February 20, 2004 Prepared for USAID/Indonesia Jl. Medan Merdeka Selatan No. 3-5 Jakarta 10110 Indonesia Prepared by Steve Rhee, M.E.Sc. Darrell Kitchener, Ph.D. Tim Brown, Ph.D. Reed Merrill, M.Sc. Russ Dilts, Ph.D. Stacey Tighe, Ph.D. Table of Contents Table of Contents............................................................................................................................. i List of Tables .................................................................................................................................. v List of Figures............................................................................................................................... vii Acronyms....................................................................................................................................... ix Executive Summary.................................................................................................................... xvii 1. Introduction............................................................................................................................1- 1 2. Legislative and Institutional Structure Affecting Biological Resources...............................2 - 1 2.1 Government of Indonesia................................................................................................2 - 2 2.1.1 Legislative Basis for Protection and Management of Biodiversity and -

Utility of a High-Resolution Mouse Single Nucleotide Polymorphism
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.29.318071; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mouse single nucleotide polymorphic targets for cross hybridization in rodents 1 2 3 4 Utility of a high-resolution mouse single nucleotide polymorphism microarray assessed for 5 rodent comparative genomics 6 7 8 9 Short title: Mouse single nucleotide polymorphic targets for cross hybridization in rodents 10 11 12 13 Rachel D. Kelly, Maja Milojevic, Freda Qi, Kathleen A. Hill* 14 15 16 17 1Department of Biology, The University of Western Ontario, London, Ontario, Canada 18 19 20 * Corresponding author 21 22 Email: [email protected] (KH) 23 24 25 26 27 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.29.318071; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mouse single nucleotide polymorphic targets for cross hybridization in rodents 28 Abstract 29 In the study of genetic diversity in non-model species there is a notable lack of the low-cost, high 30 resolution tools that are readily available for model organisms. Genotyping microarray 31 technology for model organisms is well-developed, affordable, and potentially adaptable for 32 cross-species hybridization. -

Complete Sections As Applicable
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2016.014aM officers) Short title: One (1) new species in the genus Mammarenavirus (e.g. 6 new species in the genus Zetavirus) Modules attached 2 3 4 5 (modules 1 and 11 are required) 6 7 8 9 10 Author(s): Kim Blasdell, [email protected] Veasna Duong, [email protected] Marc Eloit, [email protected] Fabrice Chretien, [email protected] Sowath Ly, [email protected] Vibol Hul, [email protected] Vincent Deubel, [email protected] Serge Morand, [email protected] Philippe Buchy, [email protected] / [email protected] Corresponding author with e-mail address: Philippe Buchy, [email protected] / [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . If in doubt, contact the appropriate subcommittee ICTV Arenaviridae Study Group chair (fungal, invertebrate, plant, prokaryote or vertebrate viruses) ICTV Study Group comments (if any) and response of the proposer: Date first submitted to ICTV: July 18, 2016 Date of this revision (if different to above): ICTV-EC comments and response of the proposer: Page 1 of 12 MODULE 2: NEW SPECIES creating and naming one or more new species. -

Μ¥ ºÉ°´˜ªržiµá¨¸Ê¥Š¨¼„—Oª¥Œ¤Äœž¦³Áš«Åš¥ A
82 ´¸¦µ¥ºÉ°´ªriµÁ¨¸Ê¥¨¼oª¥¤Ä¦³Á«Å¥ A CHECKLIST OF THE WILD MAMMALS IN THAILAND 1*/ Prateep Duengkae ABSTRACT A checklist of wild mammals (not including bats) in Thailand has been compiled based on literature surveys. The work involved the creation of a species checklist with information on locality type, species code, distribution andtaxonomic problems.The wild mammals of Thailand are comprised of 197 species, 116 genera, 28 families and 13 orders. The number of wild mammals has increased from that reported in previous studies, with some groups requiring taxonomic revision. ´¥n° ´¸¦µ¥ºÉ°´ªriµÁ¨¸Ê¥¨¼oª¥¤ (Ťn¦ª¤oµµª) ¸ÊÅo嵦¦ª¦ª¤µÁ°µ¦ ·É¡·¤¡r¨³Â®¨n o°¤¼¨ Á¥Â¡¦nµÁªÅr nµÇ 夵¦»¦ª¤¡¦o°¤´Îµ¤µÁ°Ä¦¼Â °ºÉ°Å¥ ºÉ°µ¤´ ºÉ°· ¡¦o°¤´ °´¬¦¥n° ®¨n¸É¡¦´Ê¦ µ¦Â¡¦n¦³µ¥Ä¦³Á«Å¥ ¡¦o°¤°£·¦µ¥Á¦ºÉ°°»¦¤ª·µÄµ· ¹ÉÄ {»´¦³Á«Å¥Åo¤¸¦µ¥µÅªo¨oª 197 · 116 »¨ 28 ª«r 13 °´´ ´Ê¸Ê¡ªnµÁ}¦µ¥µ·´ÊÄ®¤n (new species) °Ã¨ ¨³¦µ¥µÄ®¤n(new record) °¦³Á«Å¥ Ã¥¸É¤¸µ¨»n¤¥´o°¤¸µ¦«¹¬µÄ ¦µ¥¨³Á°¸¥Á¡·É¤¤µ ¹Êoª¥ 1£µª·µ¸ªª·¥µiµÅ¤o ³ª«µ¦r ¤®µª·¥µ¨´¥Á¬¦«µ¦r 50 ¤.Á¬¦«µ¦r .¡®¨Ã¥· ¨µ¥µª »´¦ ¦»Á¡ 10900 *E-mail:[email protected] ªµ¦µ¦´ªr µÁ¤i º°Å¥ e¸É 18 ´¸É 1 ¡.«. 2554 Journal of Wildlife in Thailand Vol.18 No.1 2011 83 εε µ¦«¹¬µÁ¡ºÉ°¦ª¦ª¤´¦µ¥¸ ºÉ°´ªrÁ¨¥¨¸Ê ¼ª¥¤Ä¦³Á«Å¥o ¸ÉÁ}Á°µ¦µ ª·µµ¦Ä´Ê nªn° e ¡.«. -

Ecologically-Based Management of Rodent Pests ECOLOGICALL V-BASED MANAGEMENT of RODENT PESTS
Ecologically-based Management of Rodent Pests ECOLOGICALL V-BASED MANAGEMENT OF RODENT PESTS Edited by: Grant R. Singleton, Lyn A. Hinds, Herwig Leirs and Zhibin Zhang Australian Centre for International Agricultural Research Canberra 1999 The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its primary mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Australian and developing country researchers in fields where Australia has special competence. Where trade names are used this constitutes neither endorsement of nor discrimination against any product by the Centre. ACIAR MONOGRAPH SERIES This peer-reviewed series contains the results of original research supported by ACIAR, or deemed relevant to ACIAR's research objectives. The series is distributed internationally, with an emphasis on the Third World ©Australian Centre for International Agricultural Research GPO Box Canberra, ACT 2601. Singleton, C.R., Hinds, L.A., Leirs, H. and Zhang, Z.ed. 1999. Ecologically-based management of rodent ACIAR Monograph No. 59, 494p. ISBN 1 86320 262 5 Editing and design by Arawang Communication Croup, Canberra Printed by Brown Prior Anderson, Melbourne, Australia page Author Contact Details 8 Abbreviations 12 List of Species 13 Preface 15 1. Ecologically-based Management of Rodent Pests-Re-evaluating 17 Our Approach to an Old Problem Grant R. Singleton, Herwig Leirs, Lyn A. Hinds and Zhibin Zhang Section 1 Basic Research - the Foundation for Sound Management 31 2. Current Paradigms of Rodent Population Dynamics- 33 What Are We Missing? Charles J. Krebs 3. The Behaviour and Ecology of Rattus norvegicus: from Opportunism to 49 Kamikaze Tendencies David W. -

Geographic Variation of Mus Caroli from East and Southeast Asia Based on Mitochondrial Cytochrome B Gene Sequences
Mammal Study 28: 67–72 (2003) © the Mammalogical Society of Japan Geographic variation of Mus caroli from East and Southeast Asia based on mitochondrial cytochrome b gene sequences Mie Terashima1, Agustinus Suyanto2, Kimiyuki Tsuchiya3, Kazuo Moriwaki4, Mei-Lei Jin5 and Hitoshi Suzuki1,* 1 Graduate School of Environmental Earth Science, Hokkaido University, Sapporo 060-0810, Japan 2 Research Centre for Biology, Indonesian Institute of Sciences, Cibinong 16911, Indonesia 3 Department of Agriculture, Tokyo University of Agriculture, Atsugi 243-0034, Japan 4 RIKEN, Bioresource Center, Tsukuba, Ibaraki 305-0074, Japan 5 Shanghai Research Center of Biotechnology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200233, China Abstract. We examined intraspecific genetic variation in the mitochondrial cytochrome b gene (1140 bp) in the Okinawa mouse, Mus caroli (Rodentia, Muridae), from Okinawa, Taiwan, Hainan, Yunnan (southern China), Thailand, Vietnam and Java, to better understand their evolutionary history and human impact on dispersal and colonization by the mice. Populations from each of the seven localities had distinctive mitochondrial DNA sequences. The genetic distances, ranging from 0.015 to 0.045 (0.033 on average), were comparable with inter-subspecies differences in the house mouse Mus musculus (0.024). Assuming that nucleotide substitutions occur at a constant rate of 0.024/million years/lineage for mitochondrial DNA, the divergence of in M. caroli is estimated to have occurred 0.3–0.9 million years ago. The relatively great extent of intraspecific mitochondrial DNA variation in this taxon, and the spatial mode of variation suggest that M. caroli is genetically structured in space, to a considerable extent. -

Isolation, Characterization, and Evolutionary Divergence of Mouse Rnase 6: Evidence for Unusual Evolution in Rodents
J Mol Evol (2004) 59:657–665 DOI: 10.1007/s00239-004-2657-0 Isolation, Characterization, and Evolutionary Divergence of Mouse RNase 6: Evidence for Unusual Evolution in Rodents Kimberly D. Dyer,1 Helene F. Rosenberg,1 Jianzhi Zhang2 1 Eosinophil Biology Section, Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA 2 Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor MI 48109, USA Received: 26 February 2004 / Accepted: 7 June 2004 [Reviewing Editor: Dr. Lauren Ancel Meyers] Abstract. The evolution of the ribonuclease A Key words: Ribonuclease — RNase A superfamily (RNase A) vertebrate-specific enzyme family is inter- — RNase k6 — RNase 6 — Host defense esting in that specific gene lineages appear to be responding to unique selective pressures in wildly diverse manners to generate proteins that are capable Introduction of reducing the infectivity of viruses, killing systemic pathogens, and inducing the growth of blood vessels all Ribonuclease 6 is a member of the ribonuclease A while maintaining the signature motifs of a ribonu- (RNase A) superfamily, which is believed to be the clease. In this paper, we present the DNA sequence and only enzyme family that is limited to vertebrates gene structure of Mus musculus RNase 6 and examine (Lander et al. 2001). This family has been well char- the expression pattern and enzymatic activity of the acterized structurally, with most members having recombinant protein. M. musculus RNase 6 has a eight cysteines as well as the lysine and the two his- limited expression pattern compared to human RNase tidines that form the catalytic triad (Beintema et al. -

List of Taxa for Which MIL Has Images
LIST OF 27 ORDERS, 163 FAMILIES, 887 GENERA, AND 2064 SPECIES IN MAMMAL IMAGES LIBRARY 31 JULY 2021 AFROSORICIDA (9 genera, 12 species) CHRYSOCHLORIDAE - golden moles 1. Amblysomus hottentotus - Hottentot Golden Mole 2. Chrysospalax villosus - Rough-haired Golden Mole 3. Eremitalpa granti - Grant’s Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus - Lowland Streaked Tenrec 3. Microgale cf. longicaudata - Lesser Long-tailed Shrew Tenrec 4. Microgale cowani - Cowan’s Shrew Tenrec 5. Microgale mergulus - Web-footed Tenrec 6. Nesogale cf. talazaci - Talazac’s Shrew Tenrec 7. Nesogale dobsoni - Dobson’s Shrew Tenrec 8. Setifer setosus - Greater Hedgehog Tenrec 9. Tenrec ecaudatus - Tailless Tenrec ARTIODACTYLA (127 genera, 308 species) ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BALAENIDAE - bowheads and right whales 1. Balaena mysticetus – Bowhead Whale 2. Eubalaena australis - Southern Right Whale 3. Eubalaena glacialis – North Atlantic Right Whale 4. Eubalaena japonica - North Pacific Right Whale BALAENOPTERIDAE -rorqual whales 1. Balaenoptera acutorostrata – Common Minke Whale 2. Balaenoptera borealis - Sei Whale 3. Balaenoptera brydei – Bryde’s Whale 4. Balaenoptera musculus - Blue Whale 5. Balaenoptera physalus - Fin Whale 6. Balaenoptera ricei - Rice’s Whale 7. Eschrichtius robustus - Gray Whale 8. Megaptera novaeangliae - Humpback Whale BOVIDAE (54 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Common Impala 3. Aepyceros petersi - Black-faced Impala 4. Alcelaphus caama - Red Hartebeest 5. Alcelaphus cokii - Kongoni (Coke’s Hartebeest) 6. Alcelaphus lelwel - Lelwel Hartebeest 7. Alcelaphus swaynei - Swayne’s Hartebeest 8. Ammelaphus australis - Southern Lesser Kudu 9. Ammelaphus imberbis - Northern Lesser Kudu 10. Ammodorcas clarkei - Dibatag 11. Ammotragus lervia - Aoudad (Barbary Sheep) 12. -

Rodent Biology and Management
Rodent Biology and Management Abstracts of papers presented at the International Conference on Rodent Biology and Management, held at Beijing, China, 5-9 October 1998 Editors: Zhi-Bin Zhang, Lyn Hinds, Grant Singleton and Zu-Wang Wang Australian Centre for International Agricultural Research Canberra 1999 The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Austral ian and developing country researchers in fields where Australia has a special research compe tence. Where trade names are used this constitutes neither endorsement of nor discrimination against any product by the Centre. ACIAR TECHNICAL REPORTS SERIES This series of publications contains technical information resulting from ACIAR-supported programs, projects, and workshops (for which proceedings are not being published), reports on Centre-supported fact-finding studies, or reports on other useful topics resulting from ACIAR activities. Publications in the series are distributed internationally to a selected audience. © Australian Centre for International Agricultural Research, GPO Box 1571. Canberra, ACT 2601. Email: [email protected] Home page: http://www.aciar.gov.au Zhi-Bin Zhang, Hinds, L., Singleton, G. and Zu-Wang Wang. ed. 1999. Rodent biology and management. Abstracts of papers presented at the International Conference on Rodent Biology and Management held -

Bonn Zoological Bulletin 63 ( 1 ): 15-114 June 2014
© Biodiversity Heritage Library, http://www.biodiversitylibrary.org/; www.zobodat.at Bonn zoological Bulletin 63 ( 1 ): 15-114 June 2014 Murine rodents (Rodentia: Murinae) of the Myanmar-Thai-Malaysian peninsula and Singapore: taxonomy, distribution, ecology, conservation status, and illustrated identification keys 12 3 12 2 3 Uraiporn Pimsai Malcolm J. Pearch Chutamas Satasook , Sara Bumrungsri & Paul J.J. Bates , , 'Princess Maha Chakri Sirindhorn Natural Histoiy Museum, Prince ofSongkla University>, Hat Yai. Songkhla Province, Thai- land 90112. E-mail: [email protected]: department ofBiology, Faculty of Science, Prince of Songkla University \ Hat Yai, Songkhla Province, Thailand 90112: sHarrison Institute, Bower-wood House, 15 St Botolph’s Road, Sevenoaks, Kent, TNI 3 3AQ, UK: E-mail: pjjbates2@hotmail. com Abstract. Based on field surveys undertaken between 2010 and 2013, museum studies in Thailand and the UK, and an extensive literature review, this paper provides information on the 28 species and 12 genera of murine rodents currently known from peninsular Myanmar, Thailand and Malaysia and Singapore. It incoiporates a detailed summary of past re- search, 1851-2013, of the Murinae in the study area and includes descriptive characters of the external, cranial and den- tal morphology and measurements for each of the rodent species. It lists and maps the 93 murine taxa described from the peninsula, 84 of which are currently considered to be synonyms at species level. Each of the 389 different localities on the 28 distribution maps is numbered and linked to its source, either literature or museum specimen, and listed in the on- line gazetteer. The global conservation status of each species is obtained from the IUCN Red List. -

Identifing Priority Ecoregions for Rodent Conservation at the Genus Level
Oryx Vol 35 No 2 April 2001 Short Communication Identifing priority ecoregions for rodent conservation at the genus level Giovanni Amori and Spartaco Gippoliti Abstract Rodents account for 40 per cent of living high number of genera) 'threat-spots' for rodent conser- mammal species. Nevertheless, despite an increased vation. A few regions, mainly drylands, are singled out interest in biodiversity conservation and their high as important areas for rodent conservation but are not species richness, Rodentia are often neglected by con- generally recognized in global biodiversity assessments. servationists. We attempt for the first time a world-wide These are the remaining forests of Togo, extreme evaluation of rodent conservation priorities at the genus 'western Sahel', the Turanian and Mongolian-Manchu- level. Given the low popularity of the order, we rian steppes and the desert of the Horn of Africa. considered it desirable to discuss identified priorities Resources for conservation must be allocated first to within the framework of established biodiversity prior- recognized threat spots and to those restricted-range ity areas of the world. Two families and 62 genera are genera which may depend on species-specific strategies recognized as threatened. Our analyses highlight the for their survival. Philippines, New Guinea, Sulawesi, the Caribbean, China temperate forests and the Atlantic Forest of Keywords Biodiversity, conservation priorities, south-eastern Brazil as the most important (for their rodents, threatened genera, world ecoregions. Conservation efforts for rodents must be included in Introduction the general framework of mammalian diversity conser- With 26-32 recognized extant families and more than vation, focusing on a biodiversity/area approach.