Isolation, Characterization, and Evolutionary Divergence of Mouse Rnase 6: Evidence for Unusual Evolution in Rodents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TAXONOMIC STUDIES from RODENT OUTBREAK AREAS in the CHITTAGONG HILL TRACTS Nikhil
Bangladesh J. Zool. 46(2): 217-230, 2018 ISSN: 0304-9027 (print) 2408-8455 (online) NEW RECORDS OF RODENT SPECIES IN BANGLADESH: TAXONOMIC STUDIES FROM RODENT OUTBREAK AREAS IN THE CHITTAGONG HILL TRACTS Nikhil Chakma*, Noor Jahan Sarker, Steven Belmain1, Sohrab Uddin Sarker, Ken Aplin2 and Sontosh Kumar Sarker3 Department of Zoology, University of Dhaka, Dhaka-1000, Bangladesh Abstract: Rodents are regarded as crop pests, significant reservoirs and vectors for many zoonotic diseases around the world. Basic taxonomic information of rodents present in a locality can help understand which species are responsible as crop pest in that habitat. The phenomenon of the 50-year cycle of gregarious bamboo flowering and rodent outbreaks in the Chittagong Hill Tracts (CHT) of Bangladesh, rodents trapping were carried out in four habitats from March, 2009 to December, 2011 in Ruma upazila of Bandarban hill district. Variety of traps were used to capture small mammals. The captured species were measured and identified using taxonomical dichotomous keys and DNA bar-coding performed in Australia. A total of 14 different small mammalian species were captured of which nine belonging to the Muridae family, and one species each of Spalacidae, Sciuridae, Tupaiidae and Soricidae families. The dominant small mammal species captured were Rattus rattus (54.06%) followed by Mus musculus (26.39%), Rattus nitidus (10.98%), Suncus murinus (5.45%), Mus terricolor (1.09%), Mus cookii nagarum (0.97%), Cannomys badius (0.16%), Leopoldamys edwardsi (0.12%), Berylmys bowersi (0.12%), Vernaya fulva (0.08%), Rattus andamanensis (0.08%), Tupaia glis (0.04%) and Callosciurus pygerythrus (0.04%). -
Red List of Endemic IUCN Red CITES Bern Bonn Georgia Species of the List Conventi Convention Caucasus on (CMS) 1
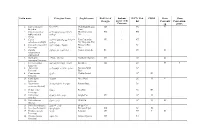
Latin name Georgian Name English name Red List of Endemic IUCN Red CITES Bern Bonn Georgia species of the list Conventi Convention Caucasus on (CMS) 1. Capra aegagrus niamori Wild Goat, Bezoar CR VU II Erxleben. Goat 2. Capra caucasica dasavleTkavkasiuri West Caucasian EN + EN Güldenstädt & jixvi Tur Pallas. 3. Capra aRmosavleTkavkasiuri East Caucasian VU + NT cylindricornis Blyth. jixvi Tur, Dagestan Tur 4. Capreolus capreolus evropuli Sveli European Roe LC Linnaeus. Deer 5. Gazella qurciki, jeirani Goitered Gazelle RE VU II subgutturosa Güldenstädt. 6. Rupicapra arCvi, fsiti Northern Chamois EN LC II rupicapra Linnaeus. 7. Cervus elaphus keTilSobili iremi Red Deer CR LC II I Linnaeus. 8. Sus scrofa gareuli Rori, taxi Eurasian Wild LC Linnaeus. Boar 9. Canis aureus tura Golden Jackal LC III Linnaeus. 10. Canis lupus mgeli Grey Wolf LC II II Linnaeus. 11. Nyctereutes enotisebri ZaRli Racoon Dog LC procyonoides Gray. 12. Vulpes vulpes mela Red Fox LC III Linnaeus. 13. Felis chaus lelianis kata Jungle Cat VU LC II Schreber. 14. Felis silvestris tyis kata Wild Cat LC II II Shreber. 15. Felis libyca Forster. velis kata Steppe Cat 16. Lynx lynx Linnaeus. focxveri Eurasian Lynx CR LC II 17. Panthera pardus jiqi Leopard CR NT I II Linnaeus. 18. Hyaena hyaena afTari Striped Hyaena CR NT Linnaeus. 19. Lutra lutra wavi Eurasian Otter, VU NT I II Linnaeus. Common Otter 20. Martes foina kldis kverna Stone Marten, LC III Erxleben. Beech Marten 21. Martes martes tyis kverna European Pine LC Linnaeus. Marten 22. Meles meles maCvi Eurasian Badger LC Linnaeus. 23. Mustela lutreola waula European Mink EN II Linnaeus. -

Morphological Comparison of Ryukyu Mouse Mus Caroli
Zoological Studies 42(2): 258-267 (2003) Morphological Comparison of Ryukyu Mouse Mus caroli (Rodentia: Muridae) Populations from Okinawajima and Taiwan Masaharu Motokawa1,*, Liang-Kong Lin2 and Junko Motokawa3 1The Kyoto University Museum, Kyoto 606-8501, Japan 2Laboratory of Wildlife Ecology, Department of Biology, Tunghai University, Taichung, Taiwan 407, R.O.C. 3Department of Zoology, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan (Accepted January 14, 2003) Masaharu Motokawa, Liang-Kong Lin and Junko Motokawa (2003) Morphological comparison of Ryukyu mouse Mus caroli (Rodentia: Muridae) populations from Okinawajima and Taiwan. Zoological Studies 42(2): 258-267. We performed univariate, bivariate, and multivariate analyses of 4 external and 17 cranial morphome- tric characters in the Ryukyu mouse, Mus caroli Bonhote, 1902 (Mammalia: Rodentia: Muridae), using 177 specimens collected from Okinawajima in the central Ryukyus and from Taiwan. There were clear morphologi- cal differences between populations from Okinawajima and Taiwan. The univariate and bivariate analyses indi- cated that the Okinawajima population differs from the Taiwan population by a shorter tail, smaller ear and audi- tory bulla, less robust incisors, larger molar row, narrower cranium in the orbital region, and longer postpalatal region. Principal component and canonical discriminant analyses based on cranial variables also suggested morphological divergence between the 2 populations. http://www.sinica.edu.tw/zool/zoolstud/42.2/258.pdf Key words: Mus caroli, M. formosanus, Taxonomy, Morphometric analyses, Allometry. The Ryukyu mouse, Mus caroli, is distrib- ognize it as a valid species (e.g., Lin and Lin uted in the Ryukyu Archipelago (Japan), Taiwan, 1983). Hainan, and southern China to the Malay Another name included in M. -

Report on Biodiversity and Tropical Forests in Indonesia
Report on Biodiversity and Tropical Forests in Indonesia Submitted in accordance with Foreign Assistance Act Sections 118/119 February 20, 2004 Prepared for USAID/Indonesia Jl. Medan Merdeka Selatan No. 3-5 Jakarta 10110 Indonesia Prepared by Steve Rhee, M.E.Sc. Darrell Kitchener, Ph.D. Tim Brown, Ph.D. Reed Merrill, M.Sc. Russ Dilts, Ph.D. Stacey Tighe, Ph.D. Table of Contents Table of Contents............................................................................................................................. i List of Tables .................................................................................................................................. v List of Figures............................................................................................................................... vii Acronyms....................................................................................................................................... ix Executive Summary.................................................................................................................... xvii 1. Introduction............................................................................................................................1- 1 2. Legislative and Institutional Structure Affecting Biological Resources...............................2 - 1 2.1 Government of Indonesia................................................................................................2 - 2 2.1.1 Legislative Basis for Protection and Management of Biodiversity and -

Utility of a High-Resolution Mouse Single Nucleotide Polymorphism
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.29.318071; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mouse single nucleotide polymorphic targets for cross hybridization in rodents 1 2 3 4 Utility of a high-resolution mouse single nucleotide polymorphism microarray assessed for 5 rodent comparative genomics 6 7 8 9 Short title: Mouse single nucleotide polymorphic targets for cross hybridization in rodents 10 11 12 13 Rachel D. Kelly, Maja Milojevic, Freda Qi, Kathleen A. Hill* 14 15 16 17 1Department of Biology, The University of Western Ontario, London, Ontario, Canada 18 19 20 * Corresponding author 21 22 Email: [email protected] (KH) 23 24 25 26 27 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.29.318071; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mouse single nucleotide polymorphic targets for cross hybridization in rodents 28 Abstract 29 In the study of genetic diversity in non-model species there is a notable lack of the low-cost, high 30 resolution tools that are readily available for model organisms. Genotyping microarray 31 technology for model organisms is well-developed, affordable, and potentially adaptable for 32 cross-species hybridization. -

Micromys Minutus)
Acta Theriol (2013) 58:101–104 DOI 10.1007/s13364-012-0102-0 SHORT COMMUNICATION The origin of Swedish and Norwegian populations of the Eurasian harvest mouse (Micromys minutus) Lars Råberg & Jon Loman & Olof Hellgren & Jeroen van der Kooij & Kjell Isaksen & Roar Solheim Received: 7 May 2012 /Accepted: 17 September 2012 /Published online: 29 September 2012 # Mammal Research Institute, Polish Academy of Sciences, Białowieża, Poland 2012 Abstract The harvest mouse (Micromys minutus) occurs the Mediterranean, from France to Russia and northwards throughout most of continental Europe. There are also two to central Finland (Mitchell-Jones et al. 1999). Recently, isolated and recently discovered populations on the it has also been found to occur in Sweden and Norway. Scandinavian peninsula, in Sweden and Norway. Here, we In 1985, an isolated population was discovered in the investigate the origin of these populations through analyses province of Dalsland in western Sweden (Loman 1986), of mitochondrial DNA. We found that the two populations and during the last decade, this population has been on the Scandinavian peninsula have different mtDNA found to extend into the surrounding provinces as well haplotypes. A comparison of our haplotypes to published as into Norway. The known distribution in Norway is sequences from most of Europe showed that all Swedish and limited to a relatively small area close to the Swedish Norwegian haplotypes are most closely related to the border, in Eidskog, Hedmark (van der Kooij et al. 2001; haplotypes in harvest mice from Denmark. Hence, the two van der Kooij et al., unpublished data). In 2007, another populations seem to represent independent colonisations but population was discovered in the province of Skåne in originate from the same geographical area. -

Complete Sections As Applicable
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2016.014aM officers) Short title: One (1) new species in the genus Mammarenavirus (e.g. 6 new species in the genus Zetavirus) Modules attached 2 3 4 5 (modules 1 and 11 are required) 6 7 8 9 10 Author(s): Kim Blasdell, [email protected] Veasna Duong, [email protected] Marc Eloit, [email protected] Fabrice Chretien, [email protected] Sowath Ly, [email protected] Vibol Hul, [email protected] Vincent Deubel, [email protected] Serge Morand, [email protected] Philippe Buchy, [email protected] / [email protected] Corresponding author with e-mail address: Philippe Buchy, [email protected] / [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . If in doubt, contact the appropriate subcommittee ICTV Arenaviridae Study Group chair (fungal, invertebrate, plant, prokaryote or vertebrate viruses) ICTV Study Group comments (if any) and response of the proposer: Date first submitted to ICTV: July 18, 2016 Date of this revision (if different to above): ICTV-EC comments and response of the proposer: Page 1 of 12 MODULE 2: NEW SPECIES creating and naming one or more new species. -

A Phylogeographic Survey of the Pygmy Mouse Mus Minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome B Sequences of Museum Specimens
A Phylogeographic Survey of the Pygmy Mouse Mus minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome b Sequences of Museum Specimens Pascale Chevret1*, Terence J. Robinson2, Julie Perez3, Fre´de´ric Veyrunes3, Janice Britton-Davidian3 1 Laboratoire de Biome´trie et Biologie Evolutive, UMR CNRS 5558, Universite´ Lyon 1, Villeurbanne, France, 2 Evolutionary Genomics Group, Department of Botany and Zoology, University of Stellenbosch, Stellenbosch, South Africa, 3 Institut des Sciences de l’Evolution de Montpellier, UMR CNRS 5554, Universite´ Montpellier 2, Montpellier, France Abstract The African pygmy mice (Mus, subgenus Nannomys) are a group of small-sized rodents that occur widely throughout sub- Saharan Africa. Chromosomal diversity within this group is extensive and numerous studies have shown the karyotype to be a useful taxonomic marker. This is pertinent to Mus minutoides populations in South Africa where two different cytotypes (2n = 34, 2n = 18) and a modification of the sex determination system (due to the presence of a Y chromosome in some females) have been recorded. This chromosomal diversity is mirrored by mitochondrial DNA sequences that unambiguously discriminate among the various pygmy mouse species and, importantly, the different M. minutoides cytotypes. However, the geographic delimitation and taxonomy of pygmy mice populations in South Africa is poorly understood. To address this, tissue samples of M. minutoides were taken and analysed from specimens housed in six South African museum collections. Partial cytochrome b sequences (400 pb) were successfully amplified from 44% of the 154 samples processed. Two species were identified: M. indutus and M. minutoides. The sequences of the M. indutus samples provided two unexpected features: i) nuclear copies of the cytochrome b gene were detected in many specimens, and ii) the range of this species was found to extend considerably further south than is presently understood. -

Mammal Survey Pol'ana (Slovakia) 2005 5�
MAMMAL SURVEY POL 'ANA (S LOVAKIA ) 2005 RESULTS OF TEN DAYS ’ FIELD WORK RAPPORT 2009.23 Juli 2009 Uitgave van de Veldwerkgroep van de Zoogdiervereniging MAMMAL SURVEY POL 'ANA (S LOVAKIA ) 2005 RESULTS OF TEN DAYS ’ FIELD WORK Editors Jan Buys Jeroen Willemsen Translations Karin de Bie Authors Jan Boshamer Jan Buys Annemarie van Diepenbeek Rob Koelman Jeroen van der Kooij Rudy van der Kuil Kees Mostert Janny Resoort Froukje Rienks Kamiel Spoelstra Anke van der Wal Jeroen Willemsen Illustration on front cover Froukje Rienks Photographs Jan Boshamer Jan Buys Jeroen van der Kooij Rudy van der Kuil Janny Resoort Kamiel Spoelstra Joost Verbeek Anke van der Wal Uitgave van de Veldwerkgroep van de Zoogdiervereniging Rapport 2009.23 Arnhem, juli 2009 ISBN: 978-90-79924-12-7 All publications of the Veldwerkgroep can be downloaded free of charge from our website: http://www.zoogdiervereniging.nl/node/137 The English page of the website contains all publications in English: http://www.zoogdiervereniging.nl/node/411 SUMMARY by: Jan Buys and Jeroen Willemsen From July 27th until August 4th 2005, the Veldwerkgroep (VWG) of the Dutch Mammal Society (Zoogdiervereniging) paid a visit to the Pol’ana Biosphere reserve in Slovakia. A workshop in the summer is a traditional part of the VWG’s yearly program, aimed at surveying mammal species which are rare or absent in the Netherlands, and to extend and exchange knowledge of survey methods and species. Secondly, these workshops abroad aim to enlarge the knowledge of presence and abundance of mammals, and to a lesser extent of other fauna groups, in the area visited. -

Revised and Commented Checklist of Mammal Species of the Romanian Fauna
REVISED AND COMMENTED CHECKLIST OF MAMMAL SPECIES OF THE ROMANIAN FAUNA DUMITRU MURARIU Abstract. Due to the permanent influences of different factors (habitat degradation andfragmentation, deforestation, infrastructure and urbanization, natural extension or decreasing of some species’ distribution, increasing number of alien species etc.), from time to time the faunistic structure of a certain area is changing. As a result of the permanent and increasing anthropic and invasive species’ pressure, our previous checklist of recent mammals from Romania (since 1984) became out of date. A number of 108 taxa are mentioned in this checklist, representing 7 orders of mammals: Insectivora (10 species), Chiroptera (30 sp.), Lagomorpha (2 sp.), Rodentia (35 sp.), Cetacea (3 sp.), Carnivora (19 sp.), Artiodactyla (8 sp.). In this list are mentioned the scientific and vernacular names (in Romanian and English languages), species distribution and conservation status, according to the Romanian regulations. Thus, only 21 species have stable populations while 76 have populations in decline or in drastic decline. Other categories are not evaluated or even present an increase in their population. Key words: species and subspecies, recent mammals, distribution, conservation. 1. INTRODUCTION MURARIU published ‘La Liste de Mammifère actuels de Roumanie; noms scientifiques et Roumains’ in 1984. That moment represented an astonishing progress in the knowledge of mammals, only in two decades. A number of 3,500 species (belonging to 1,000 genera) were reported by DAVID & GOLLEY (1965) and 4,170 species were reported by HONACKI et al. (1982). Description of new mammal species is continuing, and WILSON & REEDER DEEANN (2005) presented “... the taxonomic classification and distribution of the more than 5,400 species of mammals that exist today”. -

Seasonal Changes in Tawny Owl (Strix Aluco) Diet in an Oak Forest in Eastern Ukraine
Turkish Journal of Zoology Turk J Zool (2017) 41: 130-137 http://journals.tubitak.gov.tr/zoology/ © TÜBİTAK Research Article doi:10.3906/zoo-1509-43 Seasonal changes in Tawny Owl (Strix aluco) diet in an oak forest in Eastern Ukraine 1, 2 Yehor YATSIUK *, Yuliya FILATOVA 1 National Park “Gomilshanski Lisy”, Kharkiv region, Ukraine 2 Department of Zoology and Animal Ecology, Faculty of Biology, V.N. Karazin Kharkiv National University, Kharkiv, Ukraine Received: 22.09.2015 Accepted/Published Online: 25.04.2016 Final Version: 25.01.2017 Abstract: We analyzed seasonal changes in Tawny Owl (Strix aluco) diet in a broadleaved forest in Eastern Ukraine over 6 years (2007– 2012). Annual seasons were divided as follows: December–mid-April, April–June, July–early October, and late October–November. In total, 1648 pellets were analyzed. The most important prey was the bank vole (Myodes glareolus) (41.9%), but the yellow-necked mouse (Apodemus flavicollis) (17.8%) dominated in some seasons. According to trapping results, the bank vole was the most abundant rodent species in the study region. The most diverse diet was in late spring and early summer. Small forest mammals constituted the dominant group in all seasons, but in spring and summer their share fell due to the inclusion of birds and the common spadefoot (Pelobates fuscus). Diet was similar in late autumn, before the establishment of snow cover, and in winter. The relative representation of species associated with open spaces increased in winter, especially in years with deep snow cover, which may indicate seasonal changes in the hunting habitats of the Tawny Owl. -

Μ¥ ºÉ°´˜ªržiµá¨¸Ê¥Š¨¼„—Oª¥Œ¤Äœž¦³Áš«Åš¥ A
82 ´¸¦µ¥ºÉ°´ªriµÁ¨¸Ê¥¨¼oª¥¤Ä¦³Á«Å¥ A CHECKLIST OF THE WILD MAMMALS IN THAILAND 1*/ Prateep Duengkae ABSTRACT A checklist of wild mammals (not including bats) in Thailand has been compiled based on literature surveys. The work involved the creation of a species checklist with information on locality type, species code, distribution andtaxonomic problems.The wild mammals of Thailand are comprised of 197 species, 116 genera, 28 families and 13 orders. The number of wild mammals has increased from that reported in previous studies, with some groups requiring taxonomic revision. ´¥n° ´¸¦µ¥ºÉ°´ªriµÁ¨¸Ê¥¨¼oª¥¤ (Ťn¦ª¤oµµª) ¸ÊÅo嵦¦ª¦ª¤µÁ°µ¦ ·É¡·¤¡r¨³Â®¨n o°¤¼¨ Á¥Â¡¦nµÁªÅr nµÇ 夵¦»¦ª¤¡¦o°¤´Îµ¤µÁ°Ä¦¼Â °ºÉ°Å¥ ºÉ°µ¤´ ºÉ°· ¡¦o°¤´ °´¬¦¥n° ®¨n¸É¡¦´Ê¦ µ¦Â¡¦n¦³µ¥Ä¦³Á«Å¥ ¡¦o°¤°£·¦µ¥Á¦ºÉ°°»¦¤ª·µÄµ· ¹ÉÄ {»´¦³Á«Å¥Åo¤¸¦µ¥µÅªo¨oª 197 · 116 »¨ 28 ª«r 13 °´´ ´Ê¸Ê¡ªnµÁ}¦µ¥µ·´ÊÄ®¤n (new species) °Ã¨ ¨³¦µ¥µÄ®¤n(new record) °¦³Á«Å¥ Ã¥¸É¤¸µ¨»n¤¥´o°¤¸µ¦«¹¬µÄ ¦µ¥¨³Á°¸¥Á¡·É¤¤µ ¹Êoª¥ 1£µª·µ¸ªª·¥µiµÅ¤o ³ª«µ¦r ¤®µª·¥µ¨´¥Á¬¦«µ¦r 50 ¤.Á¬¦«µ¦r .¡®¨Ã¥· ¨µ¥µª »´¦ ¦»Á¡ 10900 *E-mail:[email protected] ªµ¦µ¦´ªr µÁ¤i º°Å¥ e¸É 18 ´¸É 1 ¡.«. 2554 Journal of Wildlife in Thailand Vol.18 No.1 2011 83 εε µ¦«¹¬µÁ¡ºÉ°¦ª¦ª¤´¦µ¥¸ ºÉ°´ªrÁ¨¥¨¸Ê ¼ª¥¤Ä¦³Á«Å¥o ¸ÉÁ}Á°µ¦µ ª·µµ¦Ä´Ê nªn° e ¡.«.